Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities
Abstract
:1. Introduction
2. Materials and Methods
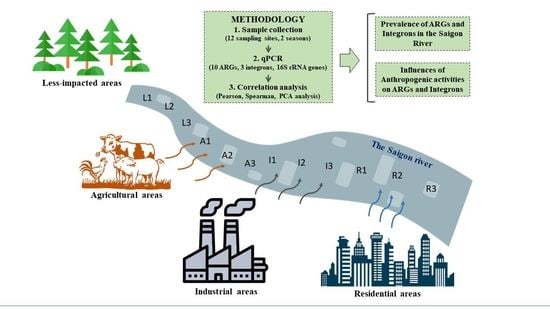
2.1. Sampling Sites
2.2. Sample Collection
2.3. DNA Extraction
2.4. Quantification of ARGs
2.5. Data Analysis
3. Results and Discussion
3.1. Occurrences of ARGs and Integrons in the Saigon River
3.2. The Abundance of ARG Genes in the Saigon River Impacted by Anthropogenic Activities
3.3. Seasonal Variation of ARGs and Integrons in the Saigon River
3.4. Correlation between the Water Quality Parameters and the Abundance of ARGs, Integrons, and 16S rRNA Gene
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jim, O. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014. [Google Scholar]
- Martin, M.J.; Thottathil, S.E.; Newman, T.B. Antibiotics Overuse in Animal Agriculture: A Call to Action for Health Care Providers. Am. J. Public Health 2015, 105, 2409–2410. [Google Scholar] [CrossRef]
- Porco, T.C.; Gao, D.; Scott, J.C.; Shim, E.; Enanoria, W.T.; Galvani, A.P.; Lietman, T.M. When Does Overuse of Antibiotics Become a Tragedy of the Commons? PLoS ONE 2012, 7, e46505. [Google Scholar] [CrossRef]
- Nguyen, H.N.K.; Van, T.T.H.; Coloe, P.J. Antibiotic resistance associated with aquaculture in Vietnam. Microbiol. Aust. 2016, 37, 108–111. [Google Scholar] [CrossRef] [Green Version]
- Kümmerer, K.; Henninger, A. Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin. Microbiol. Infect. 2003, 9, 1203–1214. [Google Scholar] [CrossRef] [Green Version]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.G.; Carmeli, Y.; Schwaber, M.J.; Chmelnitsky, I.; Schechner, V.; Navon-Venezia, S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 2010, 16, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Björkman, J.; Nagaev, I.; Berg, O.G.; Hughes, D.; Andersson, D.I. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 2000, 287, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Levin, B.R. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 1999, 2, 489–493. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, W.; Xu, T.; Zheng, B.; Yin, D. Occurrence and distribution of antibiotic resistance genes in the water and sediments of Qingcaosha Reservoir, Shanghai, China. Environ. Sci. Eur. 2019, 31, 81. [Google Scholar] [CrossRef]
- Ahammad, Z.S.; Sreekrishnan, T.R.; Hands, C.L.; Knapp, C.W.; Graham, D.W. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the upper ganges river. Environ. Sci. Technol. 2014, 48, 3014–3020. [Google Scholar] [CrossRef] [Green Version]
- Bengtsson-Palme, J.; Larsson, D.G.J. Antibiotic resistance genes in the environment: Prioritizing risks. Nat. Rev. Microbiol. 2015, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Sabri, N.A.; Schmitt, H.; Van Der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Khan, F.A.; Söderquist, B.; Jass, J. Prevalence and diversity of antibiotic resistance genes in Swedish aquatic environments impacted by household and hospital wastewater. Front. Microbiol. 2019, 10, 688. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Niu, Z.; Zhang, Y.; Zhang, K. Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them. Environ. Pollut. 2018, 236, 126–136. [Google Scholar] [CrossRef]
- Duan, M.; Gu, J.; Wang, X.; Li, Y.; Zhang, R.; Hu, T.; Zhou, B. Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotoxicol. Environ. Saf. 2019, 180, 114–122. [Google Scholar] [CrossRef]
- Chen, J.; Su, Z.; Dai, T.; Huang, B.; Mu, Q.; Zhang, Y.; Wen, D. Occurrence and distribution of antibiotic resistance genes in the sediments of the East China Sea bays. J. Environ. Sci. 2019, 81, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation Between Upstream Human Activities and Riverine Antibiotic Resistance Genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.; Berthe, T.; Budzinski, H.; Leclercq, R.; Cattoir, V.; Andremont, A.; Oberlé, K.; Laverman, A.; Denamur, E. Vulnerability and Resilience of Estuaries to Contamination by Antibiotics and Antibiotic-Resistant Bacteria: A Challenge for the Next Decade. Vulnerab. Coast. Ecosyst. Adapt. 2014, 10, 65–93. [Google Scholar]
- Czekalski, N.; Sigdel, R.; Birtel, J.; Matthews, B.; Bürgmann, H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015, 81, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Cytryn, E. Impact of anthropogenic activities on the dissemination of antibiotic resistance across ecological boundaries. Essays Biochem. 2017, 61, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kümmerer, K. Antibiotics in the aquatic environment—A review—Part I. Chemosphere 2009, 75, 417–434. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization, Pharmaceuticals in Drinking-Water; World Health Organization: Geneva, Switzerland, 2012; ISBN 9791156217343.
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human Health Risk Assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, T.-H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.-H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef]
- WEPA. WEPA Outlook on 2012 Water Environmental Management in Asia; Institute for Global Environmental Strategies: Hayame, Japan, 2012. [Google Scholar]
- World Health Organization. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization: Geneva, Switzerland, 2018; ISBN 9789241514880. [Google Scholar]
- Carrique-Mas, J.J.; Choisy, M.; Van Cuong, N.; Thwaites, G.; Baker, S. An estimation of total antimicrobial usage in humans and animals in Vietnam. Antimicrob. Resist. Infect. Control 2020, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Lulijwa, R.; Rupia, E.J.; Alfaro, A.C. Antibiotic use in aquaculture, policies and regulation, health and environmental risks: A review of the top 15 major producers. Rev. Aquac. 2020, 12, 640–663. [Google Scholar] [CrossRef]
- Nguyen Dang Giang, C.; Sebesvari, Z.; Renaud, F.; Rosendahl, I.; Hoang Minh, Q.; Amelung, W. Occurrence and Dissipation of the Antibiotics Sulfamethoxazole, Sulfadiazine, Trimethoprim, and Enrofloxacin in the Mekong Delta, Vietnam. PLoS ONE 2015, 10, e0131855. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, A.; Takada, H.; Koike, T.; Takeshita, A.; Saha, M.; Rinawati; Nakada, N.; Murata, A.; Suzuki, T.; Suzuki, S.; et al. Ubiquitous occurrence of sulfonamides in tropical Asian waters. Sci. Total Environ. 2013, 452–453, 108–115. [Google Scholar] [CrossRef]
- Hoang, T.; Vy, N.; Loan, T. Anthropogenic Input of Selected Heavy Metals (Cu, Cr, Pb, Zn and Cd) in the Aquatic Sediments of Hochiminh City, Vietnam. Water. Air. Soil Pollut. 2007, 182, 73–81. [Google Scholar] [CrossRef]
- Nguyen, T.T.N.; Némery, J.; Gratiot, N.; Strady, E.; Tran, V.Q.; Nguyen, A.T.; Aimé, J.; Peyne, A. Nutrient dynamics and eutrophication assessment in the tropical river system of Saigon—Dongnai (southern Vietnam). Sci. Total Environ. 2019, 653, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Lahens, L.; Strady, E.; Kieu-Le, T.-C.; Dris, R.; Boukerma, K.; Rinnert, E.; Gasperi, J.; Tassin, B. Macroplastic and microplastic contamination assessment of a tropical river (Saigon River, Vietnam) transversed by a developing megacity. Environ. Pollut. 2018, 236, 661–671. [Google Scholar] [CrossRef] [Green Version]
- Babut, M.; Mourier, B.; Desmet, M.; Simonnet-Laprade, C.; Labadie, P.; Budzinski, H.; De Alencastro, L.F.; Tu, T.A.; Strady, E.; Gratiot, N. Where has the pollution gone? A survey of organic contaminants in Ho Chi Minh city / Saigon River (Vietnam) bed sediments. Chemosphere 2019, 217, 261–269. [Google Scholar] [CrossRef]
- Thakali, O.; Tandukar, S.; Brooks, J.P.; Sherchan, S.P.; Sherchand, J.B.; Haramoto, E. The occurrence of antibiotic resistance genes in an Urban River in Nepal. Water (Switzerland) 2020, 12, 450. [Google Scholar] [CrossRef] [Green Version]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Su, S.; Li, C.; Yang, J.; Xu, Q.; Qiu, Z.; Xue, B.; Wang, S.; Zhao, C.; Xiao, Z.; Wang, J.; et al. Distribution of antibiotic resistance genes in three different natural water bodies-a lake, river and sea. Int. J. Environ. Res. Public Health 2020, 17, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Z.-G.; Zhang, K.; Zhang, Y. Occurrence and distribution of antibiotic resistance genes in the coastal area of the Bohai Bay, China. Mar. Pollut. Bull. 2016, 107, 245–250. [Google Scholar] [CrossRef]
- Yang, J.; Wang, H.; Roberts, D.J.; Du, H.-N.; Yu, X.-F.; Zhu, N.-Z.; Meng, X.-Z. Persistence of antibiotic resistance genes from river water to tap water in the Yangtze River Delta. Sci. Total Environ. 2020, 742, 140592. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.A.; O’Bryan, T.A.; Zurlo, J.; Miller, D.Q.; Joshi, N. Trimethoprim-Sulfamethoxazole Revisited. Arch. Intern. Med. 2003, 163, 402–410. [Google Scholar] [CrossRef]
- Kairigo, P.; Ngumba, E.; Sundberg, L.-R.; Gachanja, A.; Tuhkanen, T. Contamination of Surface Water and River Sediments by Antibiotic and Antiretroviral Drug Cocktails in Low and Middle-Income Countries: Occurrence, Risk and Mitigation Strategies. Water 2020, 12, 1376. [Google Scholar] [CrossRef]
- Thuy, H.T.T.; Nga, L.P.; Loan, T.T.C. Antibiotic contaminants in coastal wetlands from Vietnamese shrimp farming. Environ. Sci. Pollut. Res. Int. 2011, 18, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Zango, U.U.; Ibrahim, M.; Shawai, S.A.A.; Shamsuddin, I.M. A review on β-lactam antibiotic drug resistance. MOJ Drug Des. Dev. Ther. 2019, 3, 52–58. [Google Scholar]
- Graham, D.W.; Knapp, C.W.; Christensen, B.T.; McCluskey, S.; Dolfing, J. Appearance of β-lactam Resistance Genes in Agricultural Soils and Clinical Isolates over the 20th Century. Sci. Rep. 2016, 6, 21550. [Google Scholar] [CrossRef] [Green Version]
- Lenart-Boroń, A. Antimicrobial resistance and prevalence of extended-spectrum beta-lactamase genes in Escherichia coli from major rivers in Podhale, southern Poland. Int. J. Environ. Sci. Technol. 2017, 14, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Chavez, M.V.; Caicedo, L.D.; Castillo, J.E. Occurrence of β-Lactamase-Producing Gram-Negative Bacterial Isolates in Water Sources in Cali City, Colombia. Int. J. Microbiol. 2019, 2019, 1375060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.; Liu, D.; Wang, X.-H.; Wang, Y.; Zhang, B.; Wang, M.; Xu, H. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Sci. Rep. 2017, 7, 40610. [Google Scholar] [CrossRef]
- Osińska, A.; Harnisz, M.; Korzeniewska, E. Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Environ. Sci. Pollut. Res. 2016, 23, 10818–10831. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, T.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: Potential transfer to agricultural lands. Environ. Health Perspect. 2012, 120, 1144–1149. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [Green Version]
- Ullmann, I.F.; Tunsjø, H.S.; Andreassen, M.; Nielsen, K.M.; Lund, V.; Charnock, C. Detection of Aminoglycoside Resistant Bacteria in Sludge Samples From Norwegian Drinking Water Treatment Plants. Front. Microbiol. 2019, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Vakulenko, S.B.; Mobashery, S. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 2003, 16, 430–450. [Google Scholar] [CrossRef] [Green Version]
- Soler Bistué, A.J.C.; Birshan, D.; Tomaras, A.P.; Dandekar, M.; Tran, T.; Newmark, J.; Bui, D.; Gupta, N.; Hernandez, K.; Sarno, R.; et al. Klebsiella pneumoniae multiresistance plasmid pMET1: Similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS ONE 2008, 3, e1800. [Google Scholar] [CrossRef] [Green Version]
- Centrón, D.; Roy, P.H. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 2002, 46, 1402–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolmasky, M.E. Bacterial resistance to aminoglycosides and beta-lactams: The Tn1331 transposon paradigm. Front. Biosci. 2000, 5, D20–D29. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Piddock, L.J.V. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Zhang, Q. Prevalence and pollution characteristics of antibiotic resistant genes in one high anthropogenically-impacted river. PLoS ONE 2020, 15, e0231128. [Google Scholar] [CrossRef] [Green Version]
- van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [Green Version]
- Gillings, M.R. Integrons: Past, Present, and Future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef] [Green Version]
- Posada-Perlaza, C.E.; Ramírez-Rojas, A.; Porras, P.; Adu-Oppong, B.; Botero-Coy, A.-M.; Hernández, F.; Anzola, J.M.; Díaz, L.; Dantas, G.; Reyes, A.; et al. Bogotá River anthropogenic contamination alters microbial communities and promotes spread of antibiotic resistance genes. Sci. Rep. 2019, 9, 11764. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Huyan, J.; Tian, Z.; Zhang, Y.; Wen, X. Clinical class 1 integron-integrase gene—A promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant. Environ. Int. 2020, 135, 105372. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Balcázar, J.L. Real-time PCR assays for quantification of qnr genes in environmental water samples and chicken feces. Appl. Environ. Microbiol. 2013, 79, 1743–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agramont, J.; Gutierrez-Cortez, S.; Joffré, E.; Sjöling, Å.; Toledo, C.C. Antibiotic resistance genes and class 1 integron: Evidence of fecal pollution as a major driver for their abundance in water and sediments impacted by metal contamination and wastewater in the Andean region of Bolivia. bioRxiv 2020, 2020, 3350. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Le, T.-H.; Truong, T.; Hoang, T.L.; Pham, T.T.V.; Pham, T.P.T.; Tran, L.T. Influences of Anthropogenic Activities on Surface Water Quality in A Tropical River in Ho Chi Minh City, Vietnam. Int. J. Environ. Health Res. (in press).




| Human Activities | Sample Label | Description |
|---|---|---|
| Less impacted areas (L) | L1 | The site is under the Truong Phuoc Bridge, which is close to the BCR water park, which features many water recreational activities such as swimming, bathing, and water playgrounds. |
| L2 | The site is inside the Ben Duoc Temple, upstream of the Sai Gon River, which is about 5 km from industrial, agricultural, and residential areas in Ho Chi Minh City. | |
| L3 | The site is close to the Hoa Phu pump station, a water source for the Tan Hiep water treatment plant, which supplies drinking water for 11/24 districts in HCMC. | |
| Agricultural areas (A) | A1 | The site is close to the High-Tech Agricultural Park in Cu Chi District, Ho Chi Minh City. |
| A2 | The site is below the Rach Ke Bridge, which is a large canal in an agricultural area. | |
| A3 | The site is located at Binh My, a rural commune of Cu Chi District, where there are more than 20 livestock farms. | |
| Industrial areas (I) | I1 | This site is at the Xang Canal, which is an intersection of many canals passing through Tan Phu Trung (542.6 ha, main industrial fields include pharmaceuticals, food, packaging, and mechanical engineering) and Nhi Xuan (211 ha, main industrial fields include garments, food processing, plastic products, packaging, and mechanical engineering) industrial parks. |
| I2 | The site is located at Tham Luong Canal in the Tan Binh Industrial Park (128.7 ha, main industrial fields are such as packaging, cosmetic, textile, plastic product, food processing, plating and mechanical engineeing). | |
| I3 | The site is under the Suoi Cai Bridge in the Sai Gon Hi-Tech Park (913 ha, main industrial fields include microelectronics, information technology, telecommunications, precision mechanics, automation, biotechnology, pharmaceuticals, new energy, new materials, nanotechnology). | |
| Residential areas (R) | R1 | The site is located at the intersection of the Nhieu Loc-Thi Nghe Canal and Van Thanh Canal. The Nhieu Loc-Thi Nghe Canal is about 8.7 km in length and is one of the main drainage canals of Ho Chi Minh City. |
| R2 | The site is at an intersection of Te Canal, Doi Canal, Tau Hu Canal, and Ben Nghe Canal. These canals directly receive wastewater from a residential area with high population density in Ho Chi Minh City. | |
| R3 | The site is under the Cau Do Bridge, which is close to the Lang canal. This canal passes through Binh Thanh District, which is one of the most densely populated areas in Ho Chi Minh City. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truong, T.; Hoang, T.L.; Tran, L.T.; Pham, T.P.T.; Le, T.-H. Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities. Water 2021, 13, 2234. https://doi.org/10.3390/w13162234
Truong T, Hoang TL, Tran LT, Pham TPT, Le T-H. Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities. Water. 2021; 13(16):2234. https://doi.org/10.3390/w13162234
Chicago/Turabian StyleTruong, Thong, Thai Loc Hoang, Linh Thuoc Tran, Thi Phuong Thuy Pham, and Thai-Hoang Le. 2021. "Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities" Water 13, no. 16: 2234. https://doi.org/10.3390/w13162234
APA StyleTruong, T., Hoang, T. L., Tran, L. T., Pham, T. P. T., & Le, T. -H. (2021). Prevalence of Antibiotic Resistance Genes in the Saigon River Impacted by Anthropogenic Activities. Water, 13(16), 2234. https://doi.org/10.3390/w13162234