Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Diatom Sample Collection and Taxonomic Identification
2.2. Raman Spectroscopy
2.3. Data Analysis
3. Results and Discussion
3.1. Diatom Species Identification
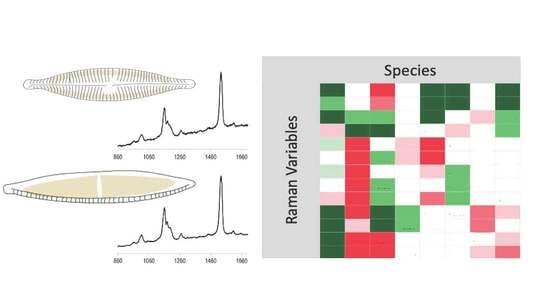
3.2. Raman Spectra
3.3. Taxonomic Identification Using Raman Data
3.3.1. Artificial Neural Network Models
3.3.2. Species Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Round, F.E.; Crawford, R.M.; Mann, D.G. Diatoms: Biology and Morphology of the Genera; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Almeida, S.F.P.; Gil, M.C.P. dÉcologie des diatomées d’eau douce de la région centrale du Portugal. Cryptogam. Algol. 2001, 22, 109–126. [Google Scholar] [CrossRef]
- Squires, L.E.; Rushforth, S.R.; Brotherson, J.D. Algal response to a thermal effluent: Study of a power station on the provo river, Utah, USA. Hydrobiologia 1979, 63, 17–32. [Google Scholar] [CrossRef]
- Vilbaste, S.; Truu, J. Distribution of benthic diatoms in relation to environmental variables in lowland streams. Hydrobiologia 2003, 493, 81–93. [Google Scholar] [CrossRef]
- Rimet, F.; Bouchez, A. Life-forms, cell-sizes and ecological guilds of diatoms in European rivers. Knowl. Manag. Aquat. Ecosyst. 2012, 406, 1. [Google Scholar] [CrossRef]
- Lear, G.; Dopheide, A.; Ancion, P.-Y.; Roberts, K.; Washington, V.; Smith, J.; Lewis, G.D. Biofilms in freshwater: Their importance for the maintenance and monitoring of freshwater health. Microb. Biofilms Curr. Res. Appl. 2012, 6700921, 129–151. [Google Scholar]
- Mendes, T.; Almeida, S.F.; Feio, M.J. Assessment of rivers using diatoms: Effect of substrate and evaluation method. Fundam. Appl. Limnol. /Arch. Für Hydrobiol. 2012, 179, 267–279. [Google Scholar] [CrossRef]
- Feio, M.J.; Hughes, R.M.; Callisto, M.; Nichols, S.J.; Odume, O.N.; Quintella, B.R.; Kuemmerlen, M.; Aguiar, F.C.; Almeida, S.F.; Alonso-EguíaLis, P. The biological assessment and rehabilitation of the world’s rivers: An overview. Water 2021, 13, 371. [Google Scholar] [CrossRef]
- Pandey, L.K.; Bergey, E.A.; Lyu, J.; Park, J.; Choi, S.; Lee, H.; Depuydt, S.; Oh, Y.-T.; Lee, S.-M.; Han, T. The use of diatoms in ecotoxicology and bioassessment: Insights, advances and challenges. Water Res. 2017, 118, 39–58. [Google Scholar] [CrossRef]
- Pinto, R.; Mortágua, A.; Almeida, S.F.; Serra, S.; Feio, M.J. Diatom size plasticity at regional and global scales. Limnetica 2020, 39, 387–403. [Google Scholar] [CrossRef]
- Keck, F.; Vasselon, V.; Tapolczai, K.; Rimet, F.; Bouchez, A. Freshwater biomonitoring in the Information Age. Front. Ecol. Environ. 2017, 15, 266–274. [Google Scholar] [CrossRef]
- Morin, S.; Gómez, N.; Tornés, E.; Licursi, M.; Rosebery, J. Benthic diatom monitoring and assessment of freshwater environments: Standard methods and future challenges. In Aquatic Biofilms: Ecology, Water Quality and Wastewater Treatment; Caister Academic Press: Norfolk, UK, 2016; p. 111. [Google Scholar]
- Alindonosi, A.; Baeshen, M.; Elsharawy, N. Prospects For Diatoms Identification Using Metagenomics: A Review. Appl. Ecol. Environ. Res. 2021, 19, 4281–4298. [Google Scholar] [CrossRef]
- Borrego-Ramos, M.; Bécares, E.; García, P.; Nistal, A.; Blanco, S. Epiphytic diatom-based biomonitoring in Mediterranean ponds: Traditional microscopy versus metabarcoding approaches. Water 2021, 13, 1351. [Google Scholar] [CrossRef]
- Coltelli, P.; Barsanti, L.; Evangelista, V.; Frassanito, A.M.; Gualtieri, P. Water monitoring: Automated and real time identification and classification of algae using digital microscopy. Environ. Sci. Processes Impacts 2014, 16, 2656–2665. [Google Scholar] [CrossRef]
- Kelly, M.; Juggins, S.; Mann, D.; Sato, S.; Glover, R.; Boonham, N.; Sapp, M.; Lewis, E.; Hany, U.; Kille, P. Development of a novel metric for evaluating diatom assemblages in rivers using DNA metabarcoding. Ecol. Indic. 2020, 118, 106725. [Google Scholar] [CrossRef]
- Kloster, M.; Langenkämper, D.; Zurowietz, M.; Beszteri, B.; Nattkemper, T.W. Deep learning-based diatom taxonomy on virtual slides. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Mora, D.; Abarca, N.; Proft, S.; Grau, J.H.; Enke, N.; Carmona, J.; Skibbe, O.; Jahn, R.; Zimmermann, J. Morphology and metabarcoding: A test with stream diatoms from Mexico highlights the complementarity of identification methods. Freshw. Sci. 2019, 38, 448–464. [Google Scholar] [CrossRef]
- Park, J.; Lee, H.; Park, C.Y.; Hasan, S.; Heo, T.-Y.; Lee, W.H. Algal morphological identification in watersheds for drinking water supply using neural architecture search for convolutional neural network. Water 2019, 11, 1338. [Google Scholar] [CrossRef] [Green Version]
- Pedraza, A.; Bueno, G.; Deniz, O.; Ruiz-Santaquiteria, J.; Sanchez, C.; Blanco, S.; Borrego-Ramos, M.; Olenici, A.; Cristobal, G. Lights and pitfalls of convolutional neural networks for diatom identification. In Optics, Photonics, and Digital Technologies for Imaging Applications V; International Society for Optics and Photonic: San Francisco, CA, USA, 2018. [Google Scholar]
- Pissaridou, P.; Vasselon, V.; Christou, A.; Chonova, T.; Papatheodoulou, A.; Drakou, K.; Tziortzis, I.; Dörflinger, G.; Rimet, F.; Bouchez, A. Cyprus’ diatom diversity and the association of environmental and anthropogenic influences for ecological assessment of rivers using DNA metabarcoding. Chemosphere 2021, 272, 129814. [Google Scholar] [CrossRef]
- Rawat, S.S.; Bisht, A.; Nijhawan, R. A Deep Learning based CNN framework approach for Plankton Classification. In Proceedings of the 2019 Fifth International Conference on Image Information Processing (ICIIP), Shimla, India, 15–17 November 2019; pp. 268–273. [Google Scholar]
- Rivera, S.; Vasselon, V.; Jacquet, S.; Bouchez, A.; Ariztegui, D.; Rimet, F. Metabarcoding of lake benthic diatoms: From structure assemblages to ecological assessment. Hydrobiologia 2018, 807, 37–51. [Google Scholar] [CrossRef]
- Rivera, S.F.; Vasselon, V.; Ballorain, K.; Carpentier, A.; Wetzel, C.E.; Ector, L.; Bouchez, A.; Rimet, F. DNA metabarcoding and microscopic analyses of sea turtles biofilms: Complementary to understand turtle behavior. PLoS ONE 2018, 13, e0195770. [Google Scholar] [CrossRef]
- Salido, J.; Sánchez, C.; Ruiz-Santaquiteria, J.; Cristóbal, G.; Blanco, S.; Bueno, G. A low-cost automated digital microscopy platform for automatic identification of diatoms. Appl. Sci. 2020, 10, 6033. [Google Scholar] [CrossRef]
- Selivanova, E.A.; Ignatenko, M.E.; Yatsenko-Stepanova, T.N.; Plotnikov, A.O. Diatom assemblages of the brackish Bolshaya Samoroda River (Russia) studied via light microscopy and DNA metabarcoding. Protistology 2019, 13, 215–235. [Google Scholar] [CrossRef]
- Zimmermann, J.; Glöckner, G.; Jahn, R.; Enke, N.; Gemeinholzer, B. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol. Ecol. Resour. 2015, 15, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Pinto, R.; Vilarinho, R.; Carvalho, A.P.; Moreira, J.A.; Guimaraes, L.; Oliva-Teles, L. Raman spectroscopy applied to diatoms (microalgae, Bacillariophyta): Prospective use in the environmental diagnosis of freshwater ecosystems. Water Res. 2021, 198, 117102. [Google Scholar] [CrossRef] [PubMed]
- Heraud, P.; Wood, B.R.; Beardall, J.; McNaughton, D. Probing the Influence of the Environment on Microalgae Using Infrared and Raman Spectroscopy. In New Approaches in Biomedical Spectroscopy; American Chemical Society: Washington, DC, USA, 2007; Volume 963, pp. 85–106. [Google Scholar]
- Parker, F.S. Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1983. [Google Scholar]
- Alexandre, M.T.; Gundermann, K.; Pascal, A.A.; van Grondelle, R.; Buchel, C.; Robert, B. Probing the carotenoid content of intact Cyclotella cells by resonance Raman spectroscopy. Photosynth. Res. 2014, 119, 273–281. [Google Scholar] [CrossRef]
- Meksiarun, P.; Spegazzini, N.; Matsui, H.; Nakajima, K.; Matsuda, Y.; Sato, H. In vivo study of lipid accumulation in the microalgae marine diatom Thalassiosira pseudonana using Raman spectroscopy. Appl. Spectrosc. 2015, 69, 45–51. [Google Scholar] [CrossRef]
- Rüger, J.; Mondol, A.S.; Schie, I.W.; Popp, J.; Krafft, C. High-throughput screening Raman microspectroscopy for assessment of drug-induced changes in diatom cells. Analyst 2019, 144, 4488–4492. [Google Scholar] [CrossRef] [Green Version]
- Pytlik, N.; Klemmed, B.; Machill, S.; Eychmüller, A.; Brunner, E. In vivo uptake of gold nanoparticles by the diatom Stephanopyxis turris. Algal Res. 2019, 39, 101447. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.; Josefson, M.; Abrahamsson, K. Characterization and mapping of carotenoids in the algae Dunaliella and Phaeodactylum using Raman and target orthogonal partial least squares. Chemom. Intell. Lab. Syst. 2011, 107, 174–177. [Google Scholar] [CrossRef]
- Wood, B.R.; Heraud, P.; Stojkovic, S.; Morrison, D.; Beardall, J.; McNaughton, D. A portable Raman acoustic levitation spectroscopic system for the identification and environmental monitoring of algal cells. Anal. Chem. 2005, 77, 4955–4961. [Google Scholar] [CrossRef]
- Yuan, P.; He, H.P.; Wu, D.Q.; Wang, D.Q.; Chen, L.J. Characterization of diatomaceous silica by Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2004, 60, 2941–2945. [Google Scholar] [CrossRef] [Green Version]
- Moreira, C.; Gomes, C.; Vasconcelos, V.; Antunes, A. Cyanotoxins occurrence in Portugal: A new report on their recent multiplication. Toxins 2020, 12, 154. [Google Scholar] [CrossRef] [Green Version]
- Saker, M.L.; Vale, M.; Kramer, D.; Vasconcelos, V.M. Molecular techniques for the early warning of toxic cyanobacteria blooms in freshwater lakes and rivers. Appl. Microbiol. Biotechnol. 2007, 75, 441–449. [Google Scholar] [CrossRef]
- Oliva-Teles, L.; Pinto, R.; Vilarinho, R.; Carvalho, A.P.; Moreira, J.A.; Guimarães, L. Environmental diagnosis with Raman Spectroscopy applied to diatoms. Biosens. Bioelectron. 2022, 198, 113800. [Google Scholar] [CrossRef]
- Lange-Bertalot, H.; Hofmann, G.; Werum, M.; Cantonati, M.; Kelly, M. Freshwater Benthic Diatoms of Central Europe: Over 800 Common Species Used in Ecological Assessment; Koeltz Botanical Books: Schmitten-Oberreifenberg, Germany, 2017; Volume 942. [Google Scholar]
- Guiry, M.D.; Guiry, G.; AlgaeBase. AlgaeBase; World-Wide Electronic Publication, National University of Ireland, Galway. Available online: https://www.algaebase.org (accessed on 20 May 2020).
- Premvardhan, L.; Bordes, L.; Beer, A.; Buchel, C.; Robert, B. Carotenoid structures and environments in trimeric and oligomeric fucoxanthin chlorophyll a/c2 proteins from resonance Raman spectroscopy. J. Phys. Chem. B 2009, 113, 12565–12574. [Google Scholar] [CrossRef]
- Oliva Teles, L.; Fernandes, M.; Amorim, J.; Vasconcelos, V. Video-tracking of zebrafish (Danio rerio) as a biological early warning system using two distinct artificial neural networks: Probabilistic neural network (PNN) and self-organizing map (SOM). Aquat. Toxicol. 2015, 165, 241–248. [Google Scholar] [CrossRef]
- Meksiarun, P.; Spegazzini, N.; Matsui, H.; Matsuda, Y.; Sato, H. Raman Spectroscopy for Monitoring CO2 Effects on Fatty Acid Synthesis of Microalgal Marine Diatom Thalassiosira pseudonana. Adv. Sci. Eng. Med. 2014, 6, 873–875. [Google Scholar] [CrossRef]
- Rüger, J.; Unger, N.; Schie, I.W.; Brunner, E.; Popp, J.; Krafft, C. Assessment of growth phases of the diatom Ditylum brightwellii by FT-IR and Raman spectroscopy. Algal Res. 2016, 19, 246–252. [Google Scholar] [CrossRef]
- Pinzaru, S.C.; Müller, C.; Tomšić, S.; Venter, M.M.; Brezestean, I.; Ljubimir, S.; Glamuzina, B. Live diatoms facing Ag nanoparticles: Surface enhanced Raman scattering of bulk cylindrotheca closterium pennate diatoms and of the single cells. RSC Adv. 2016, 6, 42899–42910. [Google Scholar] [CrossRef]
- Kuczynska, P.; Jemiola-Rzeminska, M.; Strzalka, K. Photosynthetic pigments in diatoms. Mar. Drugs 2015, 13, 5847–5881. [Google Scholar] [CrossRef]
- Novais, M.H.; Juettner, I.; Van de Vijver, B.; Morais, M.M.; Hoffmann, L.; Ector, L. Morphological variability within the Achnanthidium minutissimum species complex (Bacillariophyta): Comparison between the type material of Achnanthes minutissima and related taxa, and new freshwater Achnanthidium species from Portugal. Phytotaxa 2015, 224, 101–139. [Google Scholar] [CrossRef]
- Merlin, J.C. Resonance Raman spectroscopy of carotenoids and carotenoid-containing systems. Pure Appl. Chem. 1985, 57, 785–792. [Google Scholar] [CrossRef]
- Premvardhan, L.; Robert, B.; Beer, A.; Büchel, C. Pigment organization in fucoxanthin chlorophyll a/c2 proteins (FCP) based on resonance Raman spectroscopy and sequence analysis. Biochim. Biophys. Acta-Bioenerg. 2010, 1797, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- De Tommasi, E.; Congestri, R.; Dardano, P.; De Luca, A.C.; Managò, S.; Rea, I.; De Stefano, M. UV-shielding and wavelength conversion by centric diatom nanopatterned frustules. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Dedecker, A.P.; Goethals, P.L.; De Pauw, N. Comparison of artificial neural network (ANN) model development methods for prediction of macroinvertebrate communities in the Zwalm river basin in Flanders, Belgium. Sci. World J. 2002, 2, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Manel, S.; Dias, J.-M.; Buckton, S.; Ormerod, S. Alternative methods for predicting species distribution: An illustration with Himalayan river birds. J. Appl. Ecol. 1999, 36, 734–747. [Google Scholar] [CrossRef]
- Dedecker, A.P.; Goethals, P.L.; Gabriels, W.; De Pauw, N. Optimization of Artificial Neural Network (ANN) model design for prediction of macroinvertebrates in the Zwalm river basin (Flanders, Belgium). Ecol. Model. 2004, 174, 161–173. [Google Scholar] [CrossRef]
- Winter, M.J.; Redfern, W.S.; Hayfield, A.J.; Owen, S.F.; Valentin, J.-P.; Hutchinson, T.H. Validation of a larval zebrafish locomotor assay for assessing the seizure liability of early-stage development drugs. J. Pharmacol. Toxicol. Methods 2008, 57, 176–187. [Google Scholar] [CrossRef]
- Libreros, J.; Bueno, G.; Trujillo, M.; Ospina, M. Automated identification and classification of diatoms from water resources. In Iberoamerican Congress on Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2018; pp. 496–503. [Google Scholar]
- Lambert, D.; Green, R. Automatic Identification of Diatom Morphology using Deep Learning. In Proceedings of the 2020 35th International Conference on Image and Vision Computing New Zealand (IVCNZ), Wellington, New Zealand, 25–27 November 2020; pp. 1–7. [Google Scholar]
- Memmolo, P.; Carcagnì, P.; Bianco, V.; Merola, F.; Goncalves da Silva Junior, A.; Garcia Goncalves, L.M.; Ferraro, P.; Distante, C. Learning diatoms classification from a dry test slide by holographic microscopy. Sensors 2020, 20, 6353. [Google Scholar] [CrossRef]




| Genus | Family | Order | Subclass |
|---|---|---|---|
| Achnanthidium (72) | Achnanthidiaceae (80) | Cocconeidales (80) | Bacillariophycidae (556) |
| Planothidium (8) | |||
| Amphora (54) | Ctenulaceae (54) | Thalassiophysales (54) | |
| Cymbella (18) | Cymbelaceae (18) | Cymbellales (126) | |
| Gomphonema (108) | Gomphonemataceae (108) | ||
| Nitzschia (162) | Bacillariacea (162) | Bacillariales (162) | |
| Navicula (126) | Naviculaceae (126) | Naviculales (134) | |
| Eolimna (8) | Sellaphoraceae (8) | ||
| Fragilaria (54) | Fragilariaceae (54) | Fragillariales (72) | Fragilariophycidae (162) |
| Pseudostaurosira (18) | Staurosiraceae (18) | ||
| Ctenophora (36) | Ulnariaceae (90) | Licmophorales (90) | |
| Tabularia (36) | |||
| Ulnaria (18) | |||
| Melosira (54) | Melosiraceae (54) | Melosirales (54) | Melosirophycidae (54) |
| Cyclotella (18) | Stephanodiscaceae (18) | Stephanodiscales (18) | Thalassiosirophycidae (18) |
| Categorical Target | Species | Genus | Family | Order | Subclass |
|---|---|---|---|---|---|
| Continuous input | All | All | All | All | Width Frequency A1526NN |
| Train accuracy (%) | 49.3 | 70.1 | 74.0 | 84.2 | 78.3 |
| Test accuracy (%) | 34.9 | 52.6 | 54.9 | 58.3 | 78.9 |
| Validation accuracy (%) | 34.3 | 52.0 | 52.6 | 53.1 | 76.0 |
| Subclass | Order | Species |
|---|---|---|
| Bacillariophycidae89% | Cocconeidales 63% | Achnanthidium exiguum 67% |
| Achnanthidium minutissimum 80% | ||
| Thalassiophysales 42% | Amphora pediculus 71% | |
| Fragilariophycidae 44% | Fragilariales 47% | Fragilaria pararumpens 67% |
| Melosirophycidae 45% | Melosirales 64% | Melosira varians 82% |
| Thalassiosirophycidae 75% | Stephanodiscales 25% | Cyclotella stelligera 50% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, R.; Vilarinho, R.; Carvalho, A.P.; Moreira, J.A.; Guimarães, L.; Oliva-Teles, L. Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis. Water 2022, 14, 2116. https://doi.org/10.3390/w14132116
Pinto R, Vilarinho R, Carvalho AP, Moreira JA, Guimarães L, Oliva-Teles L. Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis. Water. 2022; 14(13):2116. https://doi.org/10.3390/w14132116
Chicago/Turabian StylePinto, Raquel, Rui Vilarinho, António Paulo Carvalho, Joaquim Agostinho Moreira, Laura Guimarães, and Luís Oliva-Teles. 2022. "Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis" Water 14, no. 13: 2116. https://doi.org/10.3390/w14132116
APA StylePinto, R., Vilarinho, R., Carvalho, A. P., Moreira, J. A., Guimarães, L., & Oliva-Teles, L. (2022). Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis. Water, 14(13), 2116. https://doi.org/10.3390/w14132116