MIMS as a Low-Impact Tool to Identify Pathogens in Water
Abstract
:1. Introduction
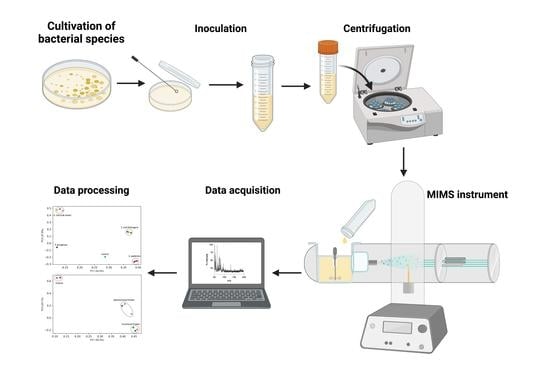
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harbor Persp. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Umber, B.J.; Shin, H.-W.; Meinardi, S.; Leu, S.-Y.; Zaldivar, F.; Cooper, D.M.; Blake, D.R. Gas Signatures from Escherichia coli and Escherichia coli-Inoculated Human Whole Blood. Clin. Transl. Med. 2013, 2, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, C.L.; Bean, H.D. Dependence of the Staphylococcal Volatilome Composition on Microbial Nutrition. Metabolites 2020, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Vilaplana, L.; Marco, M.-P. Phenazines as Potential Biomarkers of Pseudomonas Aeruginosa Infections: Synthesis Regulation, Pathogenesis and Analytical Methods for Their Detection. Anal. Bioanal. Chem. 2020, 412, 5897–5912. [Google Scholar] [CrossRef]
- Fitzgerald, S.; Holland, L.; Morrin, A. An Investigation of Stability and Species and Strain-Level Specificity in Bacterial Volatilomes. Front. Microbiol. 2021, 12, 693075. [Google Scholar] [CrossRef]
- Nizio, K.D.; Perrault, K.A.; Troobnikoff, A.N.; Ueland, M.; Shoma, S.; Iredell, J.R.; Middleton, P.G.; Forbes, S.L. In Vitro Volatile Organic Compound Profiling Using GC× GC-TOFMS to Differentiate Bacteria Associated with Lung Infections: A Proof-of-Concept Study. J. Breath Res. 2016, 10, 026008. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.; Cataneo, R.N.; Chaturvedi, A.; Kaplan, P.D.; Libardoni, M.; Mundada, M.; Patel, U.; Zhang, X. Detection of an Extended Human Volatome with Comprehensive Two-Dimensional Gas Chromatography Time-of-Flight Mass Spectrometry. PLoS ONE 2013, 8, e75274. [Google Scholar] [CrossRef]
- Heddergott, C.; Calvo, A.M.; Latgé, J.P. The Volatome of Aspergillus Fumigatus. Eukaryot. Cell 2014, 13, 1014–1025. [Google Scholar] [CrossRef] [Green Version]
- Orban, A.; Weber, A.; Herzog, R.; Hennicke, F.; Rühl, M. Transcriptome of Different Fruiting Stages in the Cultivated Mushroom Cyclocybe Aegerita Suggests a Complex Regulation of Fruiting and Reveals Enzymes Putatively Involved in Fungal Oxylipin Biosynthesis. BMC Genom. 2021, 22, 324. [Google Scholar] [CrossRef]
- Freihorst, D.; Brunsch, M.; Wirth, S.; Krause, K.; Kniemeyer, O.; Linde, J.; Kunert, M.; Boland, W.; Kothe, E. Smelling the Difference: Transcriptome, Proteome and Volatilome Changes after Mating. Fungal Genet. Biol. 2018, 112, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Boots, A.W.; Smolinska, A.; van Berkel, J.J.B.N.; Fijten, R.R.R.; Stobberingh, E.E.; Boumans, M.L.L.; Moonen, E.J.; Wouters, E.F.M.; Dallinga, J.W.; Van Schooten, F.J. Identification of Microorganisms Based on Headspace Analysis of Volatile Organic Compounds by Gas Chromatography–mass Spectrometry. J. Breath Res. 2014, 8, 027106. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of Recent Developments in GC-MS Approaches to Metabolomics-Based Research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Nalbantoglu, S. Metabolomics: Basic Principles and Strategies. In Molecular Medicine; Nalbantoglu, S., Amri, H., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Kotiaho, T.; Lauritsen, F.R.; Choudhury, T.K.; Cooks, R.G.; Tsao, G.T. Membrane Introduction Mass Spectrometry. Anal. Chem. 1991, 63, 875A–883A. [Google Scholar] [CrossRef]
- Kotiaho, T.; Lauritsen, F.R. Chapter 16 Membrane Inlet Mass Spectrometry. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2002; Volume 37, pp. 531–557. [Google Scholar]
- Giannoukos, S.; Brkić, B.; Taylor, S.; France, N. Membrane Inlet Mass Spectrometry for Homeland Security and Forensic Applications. J. Am. Soc. Mass Spectrom. 2015, 26, 231–239. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.C.; Cooks, R.G.; Allen, T.M.; Cisper, M.E.; Hemberger, P.H. Membrane Introduction Mass Spectrometry: Trends and Applications. Mass Spectrom. Rev. 2000, 19, 1–37. [Google Scholar] [CrossRef]
- Kinani, S.; Richard, B.; Souissi, Y.; Bouchonnet, S. Analysis of Inorganic Chloramines in Water. Trends Anal. Chem. 2012, 33, 55–67. [Google Scholar] [CrossRef]
- Degn, H. Membrane Inlet Mass Spectrometry in Pure and Applied Microbiology. J. Microbiol. Methods 1992, 15, 185–197. [Google Scholar] [CrossRef]
- Burlacot, A.; Burlacot, F.; Li-Beisson, Y.; Peltier, G. Membrane Inlet Mass Spectrometry: A Powerful Tool for Algal Research. Front. Plant Sci. 2020, 11, 1302. [Google Scholar] [CrossRef]
- Douchi, D.; Liang, F.; Cano, M.; Xiong, W.; Wang, B.; Maness, P.-C.; Lindblad, P.; Yu, J. Membrane-Inlet Mass Spectrometry Enables a Quantitative Understanding of Inorganic Carbon Uptake Flux and Carbon Concentrating Mechanisms in Metabolically Engineered Cyanobacteria. Front. Microbiol. 2019, 10, 1356. [Google Scholar] [CrossRef]
- Lauritsen, F.R.; Lloyd, D. Direct Detection of Volatile Metabolites Produced by Microorganisms. In Mass Spectrometry for the Characterization of Microorganisms; ACS Symposium Series; American Chemical Society: New York, NY, USA, 1993; Volume 541, pp. 91–106. ISBN 9780841227378. [Google Scholar]
- Cabral, J.P.S. Water Microbiology. Bacterial Pathogens and Water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Payment, P.; Locas, A. Pathogens in Water: Value and Limits of Correlation with Microbial Indicators. Ground Water 2011, 49, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.M.; Ismat, F.; Szakonyi, G.; Rahman, M.; Mirza, O. Probing the Putative Active Site of YjdL: An Unusual Proton-Coupled Oligopeptide Transporter from E. Coli. PLoS ONE 2012, 7, e47780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saporito, P.; Vang Mouritzen, M.; Løbner-Olesen, A.; Jenssen, H. LL-37 Fragments Have Antimicrobial Activity against Staphylococcus epidermidis Biofilms and Wound Healing Potential in HaCaT Cell Line. J. Pept. Sci. 2018, 24, e3080. [Google Scholar] [CrossRef] [Green Version]
- Saporito, P.; Biljana, M.; Løbner Olesen, A.; Jenssen, H. Antibacterial Mechanisms of GN-2 Derived Peptides and Peptoids against Escherichia coli. Biopolymers 2019, 110, e23275. [Google Scholar] [CrossRef]
- Hughes, S.S.; Nielsen, M.M.K.; Jonsbo, R.V.; Nielsen, C.U.; Lauritsen, F.R.; Prabhala, B.K. BeerMIMS: Exploring the Use of Membrane-Inlet Mass Spectrometry (MIMS) Coupled to KNIME for the Characterization of Danish Beers. Eur. J. Mass Spectrom. 2021, 27, 266–271. [Google Scholar] [CrossRef]
- Fletcher, J.S.; Vickerman, J.C. Secondary Ion Mass Spectrometry: Characterizing Complex Samples in Two and Three Dimensions. Anal. Chem. 2013, 85, 610–639. [Google Scholar] [CrossRef]
- Smith, F.T.; DeRuiter, J.; Abdel-Hay, K.; Randall Clark, C. GC–MS and FTIR Evaluation of the Six Benzoyl-Substituted-1-Pentylindoles: Isomeric Synthetic Cannabinoids. Talanta 2014, 129, 171–182. [Google Scholar] [CrossRef]
- Morris, H.R.; Taylor, G.W.; Masento, M.S.; Jermyn, K.A.; Kay, R.R. Chemical Structure of the Morphogen Differentiation Inducing Factor from Dictyostelium Discoideum. Nature 1987, 328, 811–814. [Google Scholar] [CrossRef]
- Dickschat, J.S. Capturing Volatile Natural Products by Mass Spectrometry. Nat. Prod. Rep. 2014, 31, 838–861. [Google Scholar] [CrossRef]
- Bunge, M.; Araghipour, N.; Mikoviny, T.; Dunkl, J.; Schnitzhofer, R.; Hansel, A.; Schinner, F.; Wisthaler, A.; Margesin, R.; Märk, T.D. On-Line Monitoring of Microbial Volatile Metabolites by Proton Transfer Reaction-Mass Spectrometry. Appl. Environ. Microbiol. 2008, 74, 2179–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenthal, K.; Hunsicker, E.; Ratcliffe, E.; Lindley, M.R.; Leonard, J.; Hitchens, J.R.; Turner, M.A. Volatile Atmospheric Pressure Chemical Ionisation Mass Spectrometry Headspace Analysis of E. coli and S. aureus. Anal. Methods 2021, 13, 5441–5449. [Google Scholar] [CrossRef] [PubMed]
- Key, M. A Tutorial in Displaying Mass Spectrometry-Based Proteomic Data Using Heat Maps. BMC Bioinform. 2012, 13 (Suppl. S16), S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, P.K.; Bae, H. Identification of New Metabolites of Bacterial Transformation of Indole by Gas Chromatography-Mass Spectrometry and High Performance Liquid Chromatography. Int. J. Anal. Chem. 2014, 2014, 239641. [Google Scholar] [CrossRef]
- Ratiu, I.-A.; Ligor, T.; Bocos-Bintintan, V.; Al-Suod, H.; Kowalkowski, T.; Rafińska, K.; Buszewski, B. The Effect of Growth Medium on an Escherichia Coli Pathway Mirrored into GC/MS Profiles. J. Breath Res. 2017, 11, 036012. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.; Lauritsen, F.R.; Degn, H. The Parasitic Flagellates Trichomonas Vaginalis and Tritrichomonas Foetus Produce Indole and Dimethyl Disulphide: Direct Characterization by Membrane Inlet Tandem Mass Spectrometry. J. Gen. Microbiol. 1991, 137, 1743–1747. [Google Scholar] [CrossRef] [Green Version]
- Etzkorn, J.M.; Davey, N.G.; Thompson, A.J.; Creba, A.S.; Leblanc, C.W.; Simpson, C.D.; Krogh, E.T.; Gill, C.G. The Use of MIMS-MS-MS in Field Locations as an on-Line Quantitative Environmental Monitoring Technique for Trace Contaminants in Air and Water. J. Chromatogr. Sci. 2009, 47, 57–66. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, L. A Review on Dimension Reduction. Int. Stat. Rev. 2013, 81, 134–150. [Google Scholar] [CrossRef] [Green Version]
- Simmons, S.; Peng, J.; Bienkowska, J.; Berger, B. Discovering What Dimensionality Reduction Really Tells Us About RNA-Seq Data. J. Comput. Biol. 2015, 22, 715–728. [Google Scholar] [CrossRef]
- Harder, D.; Stolz, J.; Casagrande, F.; Obrdlik, P.; Weitz, D.; Fotiadis, D.; Daniel, H. DtpB (YhiP) and DtpA (TppB, YdgR) Are Prototypical Proton-Dependent Peptide Transporters of Escherichia coli. FEBS J. 2008, 275, 3290–3298. [Google Scholar] [CrossRef]
- Prabhala, B.K.; Aduri, N.G.; Iqbal, M.; Rahman, M.; Gajhede, M.; Hansen, P.R.; Mirza, O. Several hPepT1-transported Drugs Are Substrates of the Escherichia coli Proton-Coupled Oligopeptide Transporter YdgR. Res. Microbiol. 2017, 168, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.M.; Ernst, H.A.; Wang, X.; Hald, H.; Ditta, A.C.; Ismat, F.; Rahman, M.; Mirza, O. Functional Investigation of Conserved Membrane-Embedded Glutamate Residues in the Proton-Coupled Peptide Transporter YjdL. Protein Pept. Lett. 2012, 19, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Robinson, D.K. Industrial Choices for Protein Production by Large-Scale Cell Culture. Curr. Opin. Biotechnol. 2001, 12, 180–187. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajid, S.; Aryal, I.; Chaudhri, S.F.; Lauritsen, F.R.; Jørgensen, M.G.; Jenssen, H.; Prabhala, B.K. MIMS as a Low-Impact Tool to Identify Pathogens in Water. Water 2023, 15, 184. https://doi.org/10.3390/w15010184
Sajid S, Aryal I, Chaudhri SF, Lauritsen FR, Jørgensen MG, Jenssen H, Prabhala BK. MIMS as a Low-Impact Tool to Identify Pathogens in Water. Water. 2023; 15(1):184. https://doi.org/10.3390/w15010184
Chicago/Turabian StyleSajid, Salvia, Ishika Aryal, Suleman Farooq Chaudhri, Frants Roager Lauritsen, Mikkel Girke Jørgensen, Håvard Jenssen, and Bala Krishna Prabhala. 2023. "MIMS as a Low-Impact Tool to Identify Pathogens in Water" Water 15, no. 1: 184. https://doi.org/10.3390/w15010184
APA StyleSajid, S., Aryal, I., Chaudhri, S. F., Lauritsen, F. R., Jørgensen, M. G., Jenssen, H., & Prabhala, B. K. (2023). MIMS as a Low-Impact Tool to Identify Pathogens in Water. Water, 15(1), 184. https://doi.org/10.3390/w15010184