Morphometric Differentiation of the Swimming Crab Portunus sanguinolentus (Herbst, 1783) Populations in East Asia: Implications for Stock Identification and Management
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
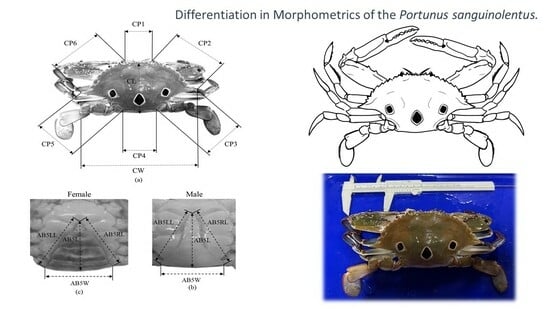
2.2. Morphometric Variation Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grimes, C.B.; Johnson, A.G.; Fable, W.A. Delineation of king mackerel (Scomberomorus cavalla) stocks along the US east coast and in the Gulf of Mexico. In Proceedings of the Stock Identification Workshop, Panama City Beach, FL, USA, 5–7 November 1985; Kumpf, H.E., Vaught, R.N., Grimes, C.B., Johnson, A.G., Nakamura, E.L., Eds.; NOAA Technical Memorandum NMFS-SEFC-199; United States Government Printing Office: Hartford, CT, USA, 1987; pp. 186–187. [Google Scholar]
- Kinsey, S.T.; Orsoy, T.; Bert, T.M.; Mahmoudi, B. Population structure of the Spanish sardine Sardinella aurita: Natural morphological variation in a genetically homogenous population. Mar. Biol. 1994, 118, 309–317. [Google Scholar] [CrossRef]
- Tzeng, C.S. Distribution of the freshwater fishes of Taiwan. J. Taiwan Mus. 1986, 39, 127–146. [Google Scholar]
- Tzeng, T.D.; Chiu, C.S.; Yeh, S.Y. Comparison of multivariate allometric coefficients in red-spot prawn (Metapenosis barbata) from adjacent waters off Taiwan. J. Taiwan Mus. 1998, 25, 85–92. [Google Scholar]
- Begg, G.A.; Friedland, K.D.; Pearce, J.B. Stock identification and its role in stock assessment and fisheries management: An overview. Fish. Res. 1999, 43, 1–8. [Google Scholar] [CrossRef]
- Tzeng, T.D.; Yeh, S.Y. Permutation tests for difference between two multivariate allometric patterns. Zool. Stud. 1999, 38, 10–18. [Google Scholar]
- Giri, F.; Collins, P.A. A geometric morphometric analysis of two sympatric species of the family Aeglidae (Crustacea, Decapoda, Anomura) from the La Plata basin. J. Zool. 2004, 71, 85–88. [Google Scholar] [CrossRef]
- Collins, P.A.; Williner, V.; Giri, F. Littoral communities: Macrocrustaceans. In The Middle Parana River, Limnology of a Subtropical Wetland; Iriondo, M.H., Paggi, J.C., Parma, M.J., Eds.; Springer: Berlin, Germany, 2007; pp. 277–302. [Google Scholar]
- Giri, F.; Loy, A. Size and shape variation of two freshwater crabs in Argentinean Patagonia: The influence of sexual dimorphism, habitat, and species interactions. J. Crustac. Biol. 2008, 28, 37–45. [Google Scholar] [CrossRef]
- Torres, M.V.; Giri, F.; Collins, P.A. Geometric morphometric analysis of the freshwater prawn Macrobrachium borellii (Decapoda: Palaemonidae) at a microgeographical scale in a floodplain system. Ecol. Res. 2014, 29, 959–968. [Google Scholar] [CrossRef]
- Pacheco, C.; Cusba, J.; Paramo, J.; Queirolo, D.; Pérez, D. Spatial structure and morphometric relationships of the deep-sea shrimp Solenocera acuminata (Decapoda, Solenoceridae) in the Colombian Caribbean. ZooKeys 2021, 1040, 1–24. [Google Scholar] [CrossRef]
- Kotlik, P.; Berrebi, P. Phylogeography of the barbel (Barbus barbus) assessed by mitochondrial DNA variation. Mol. Ecol. 2001, 10, 2177–2185. [Google Scholar] [CrossRef]
- Hellberg, M.E. Dependence of gene flow on geographic distance in two solitary corals with different larval dispersal capabilities. Evolution 1996, 50, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, T.D. Stock identification of sword prawn Parapenaeopsis hardwickii in the East China Sea and Taiwan Strait inferred by morphometric variation. Fish. Sci. 2004, 70, 758–764. [Google Scholar] [CrossRef]
- Chen, P.C.; Tzeng, T.D.; Shih, C.H.; Chu, T.J.; Lee, Y.C. Morphometric variation of the oriental river prawn (Macrobrachium nipponense) in Taiwan. Limnologica 2015, 52, 51–58. [Google Scholar] [CrossRef]
- Zelditch, M.L.; Bookstein, F.L.; Lundrigan, B.L. Ontogeny of integrated skull growth in the cotton rat Sigmodon fulviventer. Evolution 1992, 46, 1164–1180. [Google Scholar] [CrossRef]
- Thorpe, R.S.; Leamy, L. Morphometric studies in inbred and hybrid House Mice (Mus sp.): Multivariate analysis of size and shape. J. Zool. 1983, 199, 421–432. [Google Scholar] [CrossRef]
- Junquera, S.; Perez-Gandaras, G. Population diversity in Bay of Biscay anchovy (Engraulis encrasicolus L. 1785) as revealed by multivariate analysis of morphometric and meristic characters. ICES J. Mar. Sci. 1993, 50, 383–391. [Google Scholar] [CrossRef]
- Anastasiadou, C.; Liasko, R.; Leonardos, I.D. Biometric analysis of lacustrine and river inepopulations of Palemonetes antennarius (H. Milne-Edwards, 1837) (Crustacea, Decapoda, Palaemonidae) from north-western Greece. Limnologica 2009, 39, 244–254. [Google Scholar] [CrossRef]
- De Barros, M.S.F.; de Oliveira, C.D.L.; Pinto, T.K.; da Mata-Oliveira, I.; Fabré, N.N.; da Silva Batista, V. Assessment of the stock status of two penaeid shrimps in the Northeastern Brazil Marine Ecoregion and implications for their management. Reg. Stud. Mar. Sci. 2021, 48, 102046. [Google Scholar] [CrossRef]
- Strauss, R.E. Evolutionary allometry and variation in hody form in the South American catfish genus Cotydoras (Callichthyidae). Syst. Zool. 1985, 34, 381–396. [Google Scholar] [CrossRef]
- Reist, J. An empirical evaluation of several univariate methods that adjust for size variation in morphometric data. Can. J. Zool. 1985, 63, 1429–1439. [Google Scholar] [CrossRef]
- Stephenson, W.; Campbell, B. The Australian portunids (Crustacea: Portunidae). III. The genus Portunus. Mar. Freshw. Res. 1959, 10, 84–123. [Google Scholar] [CrossRef]
- Dai, A.Y.; Yang, S.L. Crabs of the China Seas; China Ocean Press: Beijing, China, 1991; pp. 4–682. [Google Scholar]
- Zafar, M.; Amin, S.M.N.; Rahman, M.M. Population dynamics of mud crab (Scylla serrata) in the Southern coastal region of Bangladesh. Asian Fish. Sci. 2006, 19, 43–50. [Google Scholar]
- Josileen, J.; Menon, N.G. Fishery and growth parameters of the blue swimmer crab Portunus pelagicus (Linnaeus, 1758) along the Mandapam coast, India. J. Mar. Biol. Assoc. India 2007, 49, 159–165. [Google Scholar]
- Sara, L.; Ingles, J.A.; Aguilar, R.O.; Laureta, L.V.; Baldevarona, R.B.; Watanabe, S. Abundance and Distribution Patterns of Scylla spp. Larvae in the Lewele Bay, Southeast Sulawesi, Indonesia. Asian Fish. Sci. 2006, 19, 331. [Google Scholar]
- Türeli, C.; Miller, T.J.; Gündogdu, S.; Yesilyurt, I.N. Growth and mortality of blue crab (Callinectes sapidus) in the North-Eastern Mediterranean Sea. J. FisheriesSciences.com 2016, 10, 55–62. [Google Scholar]
- Mantelatto, F.L.M.; Garcia, R.B. Biological aspects of the nonindigenous portunid crab Charybdis hellerii in the western tropical south Atlantic. Bull. Mar. Sci. 2001, 68, 469–477. [Google Scholar]
- Yeşilyurt, İ.N.; Türeli, C.; Gündoğdu, S. Growth parameters of the invasive blue swimming crab Portunus segnis (Forskål, 1775) (Crustacean) in the North-Eastern Mediterranean, Türkiye. Aquat. Res. 2022, 5, 285–294. [Google Scholar] [CrossRef]
- Chhapgar, B.F. On the marine crabs (Decapoda: Brachyura) of Bombay state. J. Bombay Nat. Hist. Soc. 1957, 54, 399–439. [Google Scholar]
- Sumpton, W.D.; Smith, G.S.; Potter, M.A. Notes on the biology of the portunid crab, Portunus sanguinolentus (Herbst), in subtropical queensland waters. Mar. Freshw. Res. 1989, 40, 711–717. [Google Scholar] [CrossRef]
- Campbell, G.R.; Fielder, D.R. Size at sexual maturity band occurrence of ovigerous females in three species of commercially exploited portunid crabs in S.E. Qld. Proc. R. Soc. Qld. 1986, 97, 79–87. [Google Scholar]
- Rasheed, S.; Mustaquim, J. Size at sexual maturity, breeding season and fecundity of three-spot swimming crab Portunus sanguinolentus (Herbst, 1783) (Decapoda, Brachyura, Portunidae) occurring in the coastal waters of Karachi, Pakistan. Fish. Res. 2010, 103, 56–62. [Google Scholar] [CrossRef]
- Samuel, N.J.; Soundarapandian, P. Embryonic development of commercially important portunid crab Portuns sanguinolentus (Herbst). Int. J. Anim. Vet. Adv. 2009, 1, 32–38. [Google Scholar]
- Huang, R.F. Studies on the Taxonomy and Distribution of the Portunid Crabs (Crustacea: Decapoda: Brachyura) in Taiwan. Ph.D. Dissertation, National Taiwan Ocean University, Keelung, Taiwan, 1993; p. 235. (In Chinese). [Google Scholar]
- Ye, S.Z. Fisheries biology of red spot swimming crab, Portunus sanguinolentus, on the Minnan-Taiwan bank fishing ground. Mar. Fish. 1998, 20, 60–63. (In Chinese) [Google Scholar]
- Hsu, C.C.; Chang, H.C.; Liu, H.C. Sex-variant morphometrics of the swimming crab, Portunus sanguinolentus (Herbst), from the waters off Northern Taiwan. J. Taiwan Fish. Res. 2000, 27, 175–185. [Google Scholar]
- Lee, H.H.; Hsu, C.C. Population biology of the swimming crab Portunus sanguinolentus in the waters off northern Taiwan. J. Crust. Biol. 2003, 23, 691–699. [Google Scholar] [CrossRef]
- Soundarapandian, P.; Varadharajan, D.; Boopathi, A. Reproductive biology of the commercially important portunid crab, Portunus sanguinolentus (Herbst). J. Mar. Sci. Res. Dev. 2013, 3, 2–9. [Google Scholar]
- Yang, C.P.; Li, H.X.; Li, L.; Xu, J.; Yan, Y. Population Structure, Morphometric Analysis and Reproductive Biology of Portunus Sanguinolentus (Decapoda: Brachyura: Portunidae) in Honghai Bay, South China Sea. J. Crustac. Biol. 2014, 34, 722–730. [Google Scholar] [CrossRef]
- Wimalasiri, H.B.U.G.M.; Dissanayake, D.C.T. Reproductive biology of the three-spot swimming crab (Portunus sanguinolentus) from the west coast of Sri Lanka with a novel approach to determine the maturity stage of male gonads. Invertebr. Reprod. Dev. 2016, 60, 243–253. [Google Scholar] [CrossRef]
- Wenner, A.M. Sex ratio as a function of size in marine Crustacea. Am. Nat. 1972, 106, 32–50. [Google Scholar] [CrossRef]
- Dineshbabu, A.P.; Sreedhara, B.; Muniyappa, Y. Fishery and stock assessment of Portunus sanguinolentus (Herbst) from south Karnataka coast, India. J. Mar. Biol. Assoc. India 2007, 49, 134–140. [Google Scholar]
- Sarada, P.T. Crab fishery of the Calicut coast with some aspects of the population characteristics of Portunus sanguinolentus, P. pelagicus and Charybdis cruciata. Indian J. Fish. 1998, 45, 375–386. [Google Scholar]
- Pillai, S.L.; Thirumilu, P. Fishery, biology and yield estimates of Portunus sanguinolentus off Chennai. J. Mar. Biol. Assess. 2012, 54, 73–76. [Google Scholar]
- Sumpton, W.D. Morphometric growth and fisheries biology of the crab, Charybdis natator (Herbst) in Moreton Bay, Australia (Decapoda, Brachyura). Crustaceana 1990, 59, 113–120. [Google Scholar] [CrossRef]
- Ya, Y.S. Morphometric Studies on Stock Discrimination of Swimming Crab (Charybdis feriatus) in the Offshore Areas of Taiwan. Master’s Thesis, Institute of Oceanography, National Taiwan University, Taipei, Taiwan, 2003; p. 56. (In Chinese). [Google Scholar]
- Tudela, S. Morphological variability in a Mediterranean, genetically homogeneous population of the European anchovy, Engraulis encrasicolus. Fish. Res. 1999, 42, 229–243. [Google Scholar] [CrossRef]
- Paramo, J.; Saint-Paul, U. Morphological differentiation of southern pink shrimp Farfantepenaeus notialis in Colombian Caribbean Sea. Aquat. Living Resour. 2010, 23, 95–101. [Google Scholar] [CrossRef]
- Murta, A.G. Morphological variation of horse mackerel (Trachurus trachurus) in the Iberian and North African Atlantic: Implications for stock identification. ICES J. Mar. Sci. 2000, 57, 1240–1248. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy, 1st ed.; Freeman: San Francisco, CA, USA, 1973; pp. 7–88. [Google Scholar]
- Rohlf, F.J. Tps Series; Department of Ecology and Evolution, State University of New York at Stony Brook: New York, NY, USA, 2007; pp. 7–20. [Google Scholar]
- Dryden, I.L.; Mardia, K.V. Statistical Shape Analysis; Wiley Inc.: Chichester, UK, 1998; pp. 34–56. [Google Scholar]
- Owen, J.G.; Chmielewski, M.A. On canonical variates analysis and the construction of confidence ellipses in systematic studies. Syst. Zool. 1985, 34, 366–374. [Google Scholar] [CrossRef]
- Tzeng, T.D.; Yeh, S.Y. Multivariate allometric comparisons for kuruma shrimp (Penaeus japonicus) off Taiwan. Fish. Res. 2002, 59, 279–288. [Google Scholar] [CrossRef]
- Björklund, M. Phenotypic variation of growth trajectories in finches. Evolution 1993, 47, 1506–1514. [Google Scholar] [CrossRef]
- Klingenberg, C.P.; Spence, J.R. Heterochrony and allometry: Lessons from the water strider genus Limnoporus. Evolution 1993, 47, 1834–1853. [Google Scholar] [CrossRef]
- Wiig, Ø. Multivariate variation in feral American mink (Mustela vison) from southern Norway. J. Zool. 1985, 206, 441–452. [Google Scholar] [CrossRef]
- Atchley, W.R. Some Genetic Aspects of Morphometric Variation. Numerical Taxonomy; Springer: Berlin/Heidelberg, Germany, 1983; pp. 346–363. [Google Scholar]
- Cock, A.G. Genetical aspects of metrical growth and form in animals. Q. Rev. Biophys. 1966, 41, 131–190. [Google Scholar] [CrossRef] [PubMed]
- Pepin, P.; Carr, S.M. Morphological, meristic, and genetic analysis of stock structure in juvenile Atlantic cod (Gadus morhua) from the Newfoundland shelf. Can. J. Fish. Aquat. Sci. 1993, 50, 1924–1933. [Google Scholar] [CrossRef]
- Lowe, S.A.; Van Doornik, D.M.; Winans, G.A. Geographic variation in genetic and growth patterns of Atka mackerel, Pleurogrammus monopterygius (Hexagrammidae), in the Aleutian archipelago. Fish. Bull. 1998, 96, 502–515. [Google Scholar]
- Solow, A.R. A randomization test for misclassification probability in discriminant analysis. Ecology 1990, 71, 2379–2382. [Google Scholar] [CrossRef]
- SAS. SAS User’s Guide, Statistics; SAS Institute, Inc.: Cary, NC, USA, 2014; pp. 5–33. [Google Scholar]
- Waldman, J.R.; Grossfield, J.; Wrigin, I. Review of stock discrimination techniques for striped bass. N. Am. J. Fish. Manag. 1988, 8, 410–425. [Google Scholar] [CrossRef]
- Bookstein, F.L.; Chernoff, B.; Elder, R.L.; Humphries, J.M.; Smith, G.R.; Strauss, R.E. Morphometrics in Evolutionary Biology: The Geometry of Size and Shape Change with Examples from Fishes, 15th ed.; Academy of National Sciences: Philadelphia, PA, USA, 1985; pp. 20–45. [Google Scholar]
- Sajina, A.M.; Chakraborty, S.K.; Jaiswar, A.K.; Pazhayamadam, D.G.; Sudheesan, D. Stock structure analysis of Megalaspis cordyla (Linnaeus, 1758) along the Indian coast based on truss network analysis. Fish. Res. 2011, 108, 100–105. [Google Scholar] [CrossRef]
- Anvarifar, H.; Khyabani, A.; Farahmand, H.; Vatandoust, S.; Anvarifar, H.; Jahageerdar, S. Detection of morphometric differentiation between isolated up- and downstream populations of Siah Mahi (Capoeta capoeta gracilis) (Pisces: Cyprinidae) in the Tajan River (Iran). Hydrobiologia 2011, 673, 41–52. [Google Scholar] [CrossRef]
- Wu, S.C.; Tang, H.C. Preliminary biological study of Macrobrachium nipponense. Bull. Taiwan Fish. Res. Inst. 1990, 48, 121–126. [Google Scholar]






| Area Code | Sampling Station | n | Sex | Sampling Date | CW (mm) | |
|---|---|---|---|---|---|---|
| Mean (SD) | Range | |||||
| KS | Kyushu | 58 | F | October 2021 | 102.44 (11.17) | 83.23–119.47 |
| 55 | M | 109.96 (10.99) | 78.61–127.14 | |||
| FZ | Fuzhou | 55 | F | October 2021 | 101.86 (7.88) | 89.15–122.86 |
| 45 | M | 105.91 (10.16) | 81.86–125.69 | |||
| YL | Yilan | 53 | F | January 2022 | 108.37 (11.61) | 89.15–127.97 |
| 52 | M | 116.22 (11.86) | 84.21–136.19 | |||
| XM | Xiamen | 50 | F | January 2022 | 102.67 (11.69) | 89.15–141.36 |
| 50 | M | 109.61 (10.50) | 84.21–129.30 | |||
| TN | Tainan | 53 | F | January 2022 | 113.74 (5.82) | 103.17–127.65 |
| 49 | M | 111.57 (7.35) | 95.43–131.16 | |||
| VN | Vietnam | 50 | F | January 2022 | 112.59 (9.64) | 134.92–97.23 |
| 46 | M | 109.96 (8.14) | 96.46–130.62 | |||
| HK | Hongkong | 56 | F | December 2021 | 107.72 (9.25) | 80.79–122.69 |
| 56 | M | 110.34 (13.20) | 92.49–145.48 | |||
| SG | Singapore | 55 | F | January 2022 | 119.53 (10.31) | 89.76–136.31 |
| 50 | M | 120.50 (14.30) | 100.75–158.47 | |||
| (A) Female | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | CL | CP1 | CP2 | CP3 | CP4 | CP5 | CP6 | AB3L | AB3RL | AB3LL | AB3W |
| CL | −0.848 | 0.555 | 0.728 | −0.644 | −0.786 | 0.931 | −0.253 | 0.205 ** | −0.164 ** | −0.849 ** | |
| CP1 | −0.692 ** | −0.803 ** | −0.786 ** | 0.686 ** | 0.662 ** | −0.951 | 0.52 | 0.166 ** | 0.492 ** | 0.769 ** | |
| CP2 | 0.638 ** | −0.539 ** | 0.628 ** | −0.378 ** | −0.378 ** | 0.758 | −0.443 | −0.277 ** | −0.496 ** | 0.041 | |
| CP3 | 0.780 ** | −0.514 ** | 0.766 ** | −0.092 | −0.529 ** | 0.807 | −0.394 | −0.635 ** | −0.398 ** | −0.603 ** | |
| CP4 | −0.140 ** | 0.604 ** | 0.127 ** | −0.005 | 0.682 ** | −0.66 | 0.527 | 0.231 ** | 0.496 ** | 0.724 ** | |
| CP5 | −0.136 ** | 0.553 ** | 0.231 ** | 0.201 ** | 0.773 ** | 0.021 | 0.408 | −0.706 ** | 0.333 ** | 0.858 ** | |
| CP6 | 0.929 ** | −0.745 ** | 0.834 ** | 0.862 ** | −0.114 * | −0.020 | −0.36 | −0.441 ** | −0.330 ** | −0.789 ** | |
| AB3L | 0.243 ** | 0.406 ** | 0.226 ** | 0.285 ** | 0.721 ** | 0.708 ** | 0.207 ** | 0.823 ** | 0.897 ** | 0.509 ** | |
| AB3RL | 0.541 ** | 0.136 ** | 0.365 ** | 0.501 ** | 0.558 ** | 0.562 ** | 0.474 ** | 0.917 ** | 0.866 ** | 0.081 | |
| AB3LL | 0.319 ** | 0.359 ** | 0.225 ** | 0.316 ** | 0.694 ** | 0.678 ** | 0.252 ** | 0.954 ** | 0.943 ** | 0.416 ** | |
| AB3W | −0.461 ** | 0.760 ** | −0.033 | −0.135 ** | 0.770 ** | 0.836 ** | −0.359 ** | 0.609 ** | 0.351 ** | 0.549 ** | |
| (B) Male | |||||||||||
| CL | −0.870 ** | 0.641 ** | 0.467 ** | −0.547 ** | 0.047 | 0.902 ** | −0.036 | −0.115 * | −0.753 ** | −0.907 ** | |
| CP1 | −0.730 ** | −0.742 ** | −0.555 ** | 0.577 ** | 0.631 ** | −0.949 ** | 0.463 ** | 0.051 | −0.093 | 0.862 ** | |
| CP2 | 0.742 ** | −0.482 ** | 0.524 ** | −0.299 ** | −0.259 ** | 0.767 ** | 0.261 ** | −0.102 * | 0.385 ** | −0.561 ** | |
| CP3 | 0.621 ** | −0.370 ** | 0.745 ** | −0.308 ** | −0.189 ** | 0.567 ** | −0.540 ** | 0.216 ** | 0.292 ** | −0.375 ** | |
| CP4 | −0.057 | 0.519 ** | 0.230 ** | 0.167 ** | 0.544 ** | 0.001 | 0.527 ** | 0.319 ** | 0.277 ** | 0.600 ** | |
| CP5 | −0.040 | 0.470 ** | 0.426 ** | 0.415 ** | 0.704 ** | −0.600 ** | 0.674 ** | 0.401 ** | 0.304 ** | 0.761 ** | |
| CP6 | 0.909 ** | −0.705 ** | 0.878 ** | 0.755 ** | 0.025 | 0.183 ** | −0.433 ** | −0.541 ** | 0.151 ** | −0.858 ** | |
| AB3L | −0.031 | 0.402 ** | 0.416 ** | 0.413 ** | 0.636 ** | 0.822 ** | 0.162 ** | 0.851 ** | 0.710 ** | 0.745 ** | |
| AB3RL | 0.340 ** | 0.074 | 0.683 ** | 0.636 ** | 0.549 ** | 0.750 ** | 0.511 ** | 0.905 ** | 0.860 ** | 0.391 ** | |
| AB3LL | 0.427 ** | −0.020 | 0.737 ** | 0.668 ** | 0.537 ** | 0.713 ** | 0.583 ** | 0.836 ** | 0.944 ** | 0.205 ** | |
| AB3W | −0.668 ** | 0.853 ** | −0.238 ** | −0.111 * | 0.536 ** | 0.630 ** | −0.512 ** | 0.686 ** | 0.389 ** | 0.275 ** | |
| (A) Female | ||
|---|---|---|
| Variable | First Eigenvector | Second Eigenvector |
| CL | −0.85 | 0.48 |
| CP1 | 0.94 | −0.11 |
| CP2 | −0.73 | −0.07 |
| CP3 | −0.81 | 0.15 |
| CP4 | 0.80 | 0.01 |
| CP5 | 0.79 | −0.22 |
| CP6 | −0.92 | 0.32 |
| AB3L | 0.66 | 0.69 |
| AB3RL | 0.28 | 0.94 |
| AB3LL | 0.62 | 0.75 |
| AB3W | 0.87 | −0.17 |
| Eigenvalue | 1.34 | 0.58 |
| Percentage variance | 60% | 22% |
| (B) Male | ||
| CL | −0.93 | 0.16 |
| CP1 | 0.92 | −0.27 |
| CP2 | −0.64 | 0.58 |
| CP3 | −0.50 | 0.50 |
| CP4 | 0.70 | 0.18 |
| CP5 | 0.79 | 0.27 |
| CP6 | −0.91 | 0.32 |
| AB3L | 0.70 | 0.67 |
| AB3RL | 0.31 | 0.90 |
| AB3LL | 0.15 | 0.92 |
| AB3W | 0.96 | 0.09 |
| Eigenvalue | 3.18 | 0.42 |
| Percentage variance | 53% | 27% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, W.-C.; Chang, P.-H.; Shih, C.-H.; Chen, P.-C.; Tzeng, T.-D.; Han, Y.-S.; Lu, Y.-M. Morphometric Differentiation of the Swimming Crab Portunus sanguinolentus (Herbst, 1783) Populations in East Asia: Implications for Stock Identification and Management. Water 2023, 15, 3335. https://doi.org/10.3390/w15193335
Kao W-C, Chang P-H, Shih C-H, Chen P-C, Tzeng T-D, Han Y-S, Lu Y-M. Morphometric Differentiation of the Swimming Crab Portunus sanguinolentus (Herbst, 1783) Populations in East Asia: Implications for Stock Identification and Management. Water. 2023; 15(19):3335. https://doi.org/10.3390/w15193335
Chicago/Turabian StyleKao, Wei-Chieh, Ping-Hung Chang, Chun-Han Shih, Po-Cheng Chen, Tzong-Der Tzeng, Yu-San Han, and Yu-Ming Lu. 2023. "Morphometric Differentiation of the Swimming Crab Portunus sanguinolentus (Herbst, 1783) Populations in East Asia: Implications for Stock Identification and Management" Water 15, no. 19: 3335. https://doi.org/10.3390/w15193335
APA StyleKao, W. -C., Chang, P. -H., Shih, C. -H., Chen, P. -C., Tzeng, T. -D., Han, Y. -S., & Lu, Y. -M. (2023). Morphometric Differentiation of the Swimming Crab Portunus sanguinolentus (Herbst, 1783) Populations in East Asia: Implications for Stock Identification and Management. Water, 15(19), 3335. https://doi.org/10.3390/w15193335