Microbial Community Structure of a Leachfield Soil: Response to Intermittent Aeration and Tetracycline Addition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Facility

2.2. Aeration
2.3. Antibiotic Dosing
2.4. Soil Sampling
2.5. Phospholipid Fatty Acid Analysis
2.6. DNA Extraction from Soil
2.7. PCR-DGGE
2.8. Clone Libraries
2.9. Data Analysis
3. Results
3.1. Effects of Intermittent Aeration
3.1.1. PLFA Analysis
| Tmt | Day | Total PLFA concentration a (nmol g−1 soil) | Community structure | |||||
|---|---|---|---|---|---|---|---|---|
| Firmicutes/Anaerobic G− bacteria | Proteobacteria | Anaerobic metal reducers | Actinomycetes/SRB | General | Eukaryotes | |||
| ————————— % of total PLFA —————————— | ||||||||
| AIR | 0 | 117,673 | 9.3 | 63.5 | 2.4 | 0.6 | 21.1 | 3.1 |
| 11 | 55,305 | 8.5 | 61.2 | 2.4 | 0.8 | 21.5 | 5.5 | |
| LEACH | 0 | 58,599 | 13.3 | 54.2 | 1.9 | 1.3 | 26.8 | 2.6 |
| 11 | 53,819 | 11.9 | 55.6 | 1.9 | 1.3 | 26.5 | 2.9 | |

3.1.2. PCR-DGGE Analysis
| Treatment | Day 0 | Day 11 |
|---|---|---|
| AIR | 50.7 | 44.0 |
| LEACH | 27.0 | 23.0 |
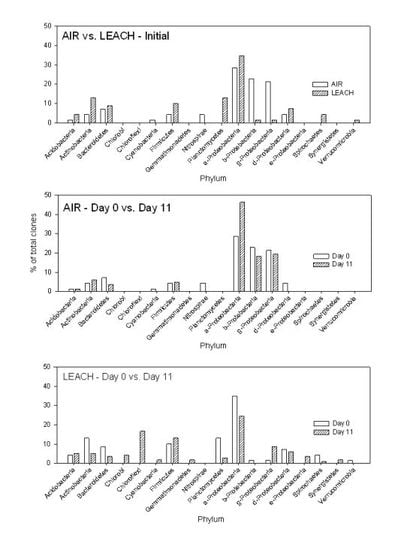
3.1.3. Clone Libraries

| Phylum | Genus and species | Treatment | Potential function | |||
|---|---|---|---|---|---|---|
| AIR | LEACH | |||||
| Day 0 | Day 11 | Day 0 | Day 11 | |||
| Acidobacteria | Terriglobus roseus | Extracellular polysaccharide production [28] | ||||
| Actinobacteria | Leucobacter komagatae | Biosurfactant production [29] | ||||
| Mycobacterium arupense | Pathogen [30] | |||||
| Mycobacterium sp. | Pathogen; PAH degradation [30,31] | |||||
| Rhodococcus coprophilus | Phenol degradation [32] | |||||
| Bacteroidetes | Flavobacterium succinicans | X | Cellulose & polysaccharide degradation [33] | |||
| Firmicutes | Bacillus sp. | Pathogen; various | ||||
| Clostridium sp. | Pathogen; various | |||||
| Nitrospirae | Nitrospira sp. | X | NO2− oxidation [34] | |||
| α-Proteobacteria | Caulobacter sp. | X | Unknown [35] | |||
| Phenylobacterium sp. | X | Degradation of chlorinated N-heterocyclics & linear alkylbenzenesulfonates [36] | ||||
| Beijerinckia sp. | X | Non-symbiotic N fixation; degradation of aromatic compounds [37] | ||||
| Afipia sp. | Pathogen [38] | |||||
| Bradyrhizobium elkanii | Symbiotic N fixation [39] | |||||
| Nitrobacter vulgaris | NO2− oxidation [40] | |||||
| Methylocystis parvus | X | CH4 oxidation [41] | ||||
| Methylocystis sp. | X | CH4 oxidation [41] | ||||
| Labrys sp. | X | Unknown | ||||
| Erythrobacter sp. | Aerobic phototrophic bacteria | |||||
| Sphingobium sp. | X | Degradation of phenolic compounds [42] | ||||
| Sphingopyxis sp. | X | Degradation of polyvinyl alcohols [42] | ||||
| β-Proteobacteria | Acidovorax defluvii | X | Denitrification [43] | |||
| Acidovorax facilis | X | Degradation of polyhydroxyalkanoates [44] | ||||
| Thiobacillus sp. | X | Fe, S & S2− oxidation | ||||
| Dechloromonas sp. | Perchlorate reduction [45] | |||||
| Rhodocyclus tenuis | Purple, non-S photosynthetic bacteria; methanol & formate oxidation | |||||
| Zoogloea ramigera | X | Extracellular polysaccharide production | ||||
| δ-Proteobacteria | Desulfovibrio desulfuricans | X | SO42− & NO3− reduction | |||
| γ-Proteobacteria | Legionella pneumophila | X | Pathogen [46] | |||
| Methylosarcina sp. | X | Methane oxidation [47] | ||||
| Pseudomonas stutzeri | X | Pathogen; denitrification; degradation of CCl4[48,49,50] | ||||
| Pseudomonas umsongensis | X | Various [51] | ||||
| Pseudomonas sp. | X | Various | ||||
| Luteibacter rhizovicinus | X | X | Chitin degradation [52] | |||
| Lysobacter sp. | X | Glucan & chitin degradation [53] | ||||
| Thermomonas sp. | Fe2+ oxidation; NO3− reduction [54] | |||||
3.2. Effects of Tetracycline
3.2.1. PLFA Analysis
3.2.2. PCR-DGGE Analysis
3.2.3. Clone Libraries
4. Discussion
4.1. Effects of Intermittent Aeration
4.2. Effects of Tetracycline
5. Conclusions
Acknowledgments
References
- Sanz, J.L.; Kochling, T. Molecular biology techniques used in wastewater treatment: An overview. Process Biochem. 2007, 42, 119–133. [Google Scholar] [CrossRef]
- Calaway, W.T.; Carroll, W.R.; Long, S.K. Heterotrophic bacteria encountered in intermittent sand filtration of sewage. Sew. Ind. Wastes 1952, 24, 642–653. [Google Scholar]
- Pell, M.; Nyberg, F. Infiltration of wastewater in a newly started pilot sand-filter system: II. Development and distribution of the bacterial populations. J. Environ. Qual. 1989, 18, 457–462. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Amador, J.A.; Potts, D.A.; Savin, M.C.; Tomlinson, P.; Görres, J.H.; Nicosia, E.L. Mesocosm-scale evaluation of faunal and microbial communities of aerated and unaerated leachfield soil. J. Environ. Qual. 2006, 35, 1160–1169. [Google Scholar] [CrossRef]
- Potts, D.A.; Görres, J.H.; Nicosia, E.L.; Amador, J.A. Effects of aeration on water quality from septic system leachfields. J. Environ. Qual. 2004, 33, 1828–1838. [Google Scholar] [CrossRef]
- Tomaras, J.; Sahl, J.W.; Siegrist, R.L.; Spear, J.R. Microbial diversity of septic tank effluent and a soil biomat. Appl. Environ. Microbiol. 2009, 75, 3348–3351. [Google Scholar] [CrossRef]
- Rabølle, M.; Spliid, N.H. Sorption and mobility of metronidazole, olaquindox, oxytetracycline, and tylosin in soil. Chemosphere 2000, 40, 715–722. [Google Scholar] [CrossRef]
- Loke, M.J.T.; Halling-Sørensen, B. Determination of the distribution coefficient (logKd) of oxytetracycline, tylosin A, olaquindox and metronidazole in manure. Chemosphere 2002, 48, 351–361. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Halling-Sørensen, B.; Sengelov, G.; Tjornelund, J. Toxicity of tetracyclines and tetracycline degradation products to environmentally-relevant bacteria, including selected tetracycline-resistant bacteria. Arch. Environ. Contam. Toxicol. 2002, 42, 263–271. [Google Scholar]
- Amador, J.A.; Potts, D.A.; Loomis, G.W.; Kalen, D.V.; Patenaude, E.L.; Görres, J.H. Improvement of hydraulic and water quality renovation functions by intermittent aeration of soil treatment areas in onsite wastewater treatment systems. Water 2010, 2, 886–903. [Google Scholar]
- Patenaude, E.L.; Atoyan, J.A.; Potts, D.A.; Amador, J.A. Effects of tetracycline on water quality, soil and gases in aerated and unaerated leachfield mesocosms. J. Environ. Sci. Health Pt. A 2008, 43, 1054–1063. [Google Scholar] [CrossRef]
- Atoyan, J.A.; Patenaude, E.L.; Potts, D.A.; Amador, J.A. Effects of tetracycline on antibiotic resistance and removal of fecal indicator bacteria in aerated and unaerated leachfield mesocosms. J. Environ. Sci. Health Part A 2007, 42, 1571–1578. [Google Scholar] [CrossRef]
- White, D.C.; Davis, W.M.; Nickels, J.S.; King, J.D.; Bobbie, R.J. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 1979, 40, 51–62. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Tunlid, A.; Hoitink, J.A.; Low, C.; White, D.C. Characterization of bacteria that suppress Rhizoctonia damping-off in bark compost media by analysis of fatty acid biomarkers. Appl. Environ. Microbiol. 1989, 55, 1368–1374. [Google Scholar]
- Dowling, N.J.E.; Widdel, F.; White, D.C. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulfate reducers and other sulfide forming bacteria. J. Gen. Microbiol. 1986, 132, 1815–1825. [Google Scholar]
- Edlund, A.; Nichols, P.D.; Roffey, R.; White, D.C. Extractable and lipopolysaccharide fatty acid and hydroxyl acid profiles from Desulfovibrio species. J. Lipid Res. 1985, 26, 982–988. [Google Scholar]
- White, D.C.; Pinkart, H.C.; Ringelberg, D.B. Biomass measurements: Biochemical approaches. In Manual of Environmental Microbiology; Hurst, C.J., Knudsen, G.R., McInerney, M.J., Stetzenbach, L.D., Walter, M.V., Eds.; ASM Press: Washington, DC, USA, 1997; pp. 91–101. [Google Scholar]
- White, D.C.; Stair, J.O.; Ringelberg, D.B. Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Ind. Microbiol. 1996, 17, 185–196. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar]
- Ferris, M.J.; Muyzer, G.; Ward, D.M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar]
- Rasband, W.S. ImageJ, Version 1.38; U.S National Institutes of Health: Bethesda, MD, USA, 1997–2007. Available online: http://rsb.info.nih.gov/ij/(accessed on 16 April 2013).
- DeSantis, T.Z.; Hugenholtz, P.; Keller, K.; Brodie, E.L.; Larsen, N.; Piceno, Y.M.; Phan, R.; Andersen, G.L. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006, 34, W394–W399. [Google Scholar] [CrossRef]
- Staddon, W.J.; Duchesne, L.C.; Trevors, J.T. Microbial diversity and community structure of postdisturbance forest soils as determined by sole-carbon-source utilization patterns. Microb. Ecol. 1997, 34, 125–130. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Breznak, J.A.; Schmidt, T.M. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the Phylum Acidobacteria. Appl. Environ. Microbiol. 2007, 73, 2708–2717. [Google Scholar] [CrossRef]
- Takeuchi, M.; Weiss, N.; Schumann, P.; Yokota, A. Leucobacter komagatae gen. nov., sp. nov., a new aerobic Gram-positive, nonsporulating rod with 2,4-diaminobutyric acid in the cell wall. Int. J. Syst. Bacteriol. 1996, 46, 967–971. [Google Scholar]
- Tortoli, E. The new mycobacteria: An update. FEMS Immunol. Med. Microbiol. 2006, 48, 159–178. [Google Scholar] [CrossRef]
- Leys, N.M.; Ryngaert, A.; Bastiaens, L.; Wattiau, P.; Top, E.M.; Verstraete, W.; Springael, D. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 2005, 51, 375–388. [Google Scholar]
- Mara, D.D.; Oragui, J.I. Occurrence of Rhodococcus coprophilus and associated actinomycetes in feces, sewage, and freshwater. Appl. Environ. Microbiol. 1981, 42, 1037–1042. [Google Scholar]
- Anderson, R.L.; Ordal, E.J. Cytophaga succinicans sp. n., a facultatively anaerobic, aquatic myxobacterium. J. Bacteriol. 1961, 81, 130–138. [Google Scholar]
- Koops, H.-P.; Pommerening-Röser, A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 2001, 37, 1–9. [Google Scholar] [CrossRef]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.; Heidelberg, J.F.; Alley, M.R.K.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef]
- Ke, N.; Xiao, C.; Ying, Q.; Ji, S. A new species of the genus Phenylobacterium for the degradation of LAS (linear alkylbenzene sulfonate). [in Chinese]. Wei Sheng Wu Xue Bao 2003, 43, 1–7. [Google Scholar]
- Gibson, D.T. Beijerinckia sp strain B1: A strain by any other name. J. Ind. Microbiol. 1999, 23, 284–293. [Google Scholar] [CrossRef]
- Brenner, D.J.; Hollis, D.G.; Moss, C.W.; English, C.K.; Hall, G.S.; Vincent, J.; Radosevic, J.; Birkness, K.A.; Bibb, W.F.; Quinn, F.D. Proposal of Afipia gen. nov., with Afipia felis sp. nov. (formerly the cat scratch disease bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland Clinic Foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J. Clin. Microbiol. 1991, 29, 2450–2460. [Google Scholar]
- Rumjanek, N.G.; Dobert, R.C.; van Berkum, P.; Triplett, E.W. Common soybean inoculant strains in Brazil are members of Bradyrhizobium elkanii. Appl. Environ. Microbiol. 1993, 59, 4371–4373. [Google Scholar]
- Bock, E.; Koops, H.-P.; Möller, U.C.; Rudert, M. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch. Microbiol. 1990, 153, 105–110. [Google Scholar] [CrossRef]
- Hou, C.T.; Laskin, A.I.; Patel, R.N. Growth and polysaccharide production by Methylocystis parvus OBBP on methanol. Appl. Environ. Microbiol. 1979, 37, 800–804. [Google Scholar]
- Takeuchi, M.; Hamana, K.; Hiraishi, A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [Google Scholar]
- Schulze, R.; Spring, S.; Amann, R.; Huber, I.; Ludwig, W.; Schleifer, K.H.; Kämpfer, P. Genotypic diversity of Acidovorax strains isolated from activated sludge and description of Acidovorax defluvii sp. nov. Syst. Appl. Microbiol. 1999, 22, 205–214. [Google Scholar] [CrossRef]
- Mergaert, J.; Webb, A.; Anderson, C.; Wouters, A.; Swings, J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl. Environ. Microbiol. 1993, 59, 3233–3238. [Google Scholar]
- Achenbach, L.A.; Michaelidou, U.; Bruce, R.A.; Fryman, J.; Coates, J.D. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 2001, 51, 527–533. [Google Scholar]
- Friedman, H.; Yamamoto, Y.; Klein, T.W. Legionella pneumophila pathogenesis and immunity. Sem. Pediatr. Infect. Dis. 2002, 13, 273–279. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Stolyar, S.M.; Auman, A.J.; Lara, J.C.; Lidstrom, M.E.; Chistoserdova, L. Methylosarcina lacus sp. nov., a methanotroph from Lake Washington, Seattle, USA, and emended description of the genus Methylosarcina. Int. J. Syst. Evol. Microbiol. 2005, 55, 2345–2350. [Google Scholar] [CrossRef]
- Lalucat, J.; Bennasar, A.; Bosch, R.; García-Valdés, E.; Palleroni, N.J. Biology of Pseudomonas stutzeri. Microbiol. Molec. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef]
- Baggi, G.; Barbieri, P.; Galli, E.; Tollari, S. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl. Environ. Microbiol. 1987, 53, 2129–2132. [Google Scholar]
- Zumft, W.G. The denitrifying prokaryotes. In The Prokaryotes; Balows, A., Ed.; Springer-Verlag: New York, NY, USA, 1992. [Google Scholar]
- Kwon, S.W.; Kim, J.S.; Park, I.C.; Yoon, S.H.; Park, D.H.; Lim, C.K.; Go, S.J. Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel species from farm soils in Korea. Int. J. Syst. Evol. Microbiol. 2003, 53, 21–27. [Google Scholar] [CrossRef]
- Johansen, J.E.; Binnerup, S.J.; Kroer, N.; Mølbak, L. Luteibacter rhizovicinus gen. nov., sp. nov., a yellow-pigmented gammaproteobacterium isolated from the rhizosphere of barley (Hordeum vulgare L.). Int. J. Syst. Evol. Microbiol. 2008, 55, 2285–2291. [Google Scholar]
- Christensen, P.; Cook, F.D. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bacteriol. 1978, 28, 367–393. [Google Scholar] [CrossRef]
- Mergaert, J.; Cnockaert, M.C.; Swings, J. Thermomonas fusca sp. nov. and Thermomonas brevis sp. nov., two mesophilic species isolated from a denitrification reactor with poly(epsilon-caprolactone) plastic granules as fixed bed, and emended description of the genus Thermomonas. Int. J. Syst. Evol. Microbiol. 2003, 53, 1961–1966. [Google Scholar] [CrossRef]
- Atlas, R.M.; Bartha, R. Microbial Ecology: Fundamentals and Applications, 4th ed; Benjamin Cummings: Menlo Park, CA, USA, 1998. [Google Scholar]
- Dantas, G.; Sommer, M.O.A.; Oluwasegun, R.D.; Church, G.M. Bacteria subsisting on antibiotics. Science 2008, 320, 100–103. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Atoyan, J.A.; Staroscik, A.M.; Nelson, D.R.; Patenaude, E.L.; Potts, D.A.; Amador, J.A. Microbial Community Structure of a Leachfield Soil: Response to Intermittent Aeration and Tetracycline Addition. Water 2013, 5, 505-524. https://doi.org/10.3390/w5020505
Atoyan JA, Staroscik AM, Nelson DR, Patenaude EL, Potts DA, Amador JA. Microbial Community Structure of a Leachfield Soil: Response to Intermittent Aeration and Tetracycline Addition. Water. 2013; 5(2):505-524. https://doi.org/10.3390/w5020505
Chicago/Turabian StyleAtoyan, Janet A., Andrew M. Staroscik, David R. Nelson, Erika L. Patenaude, David A. Potts, and José A. Amador. 2013. "Microbial Community Structure of a Leachfield Soil: Response to Intermittent Aeration and Tetracycline Addition" Water 5, no. 2: 505-524. https://doi.org/10.3390/w5020505
APA StyleAtoyan, J. A., Staroscik, A. M., Nelson, D. R., Patenaude, E. L., Potts, D. A., & Amador, J. A. (2013). Microbial Community Structure of a Leachfield Soil: Response to Intermittent Aeration and Tetracycline Addition. Water, 5(2), 505-524. https://doi.org/10.3390/w5020505