Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Recombinant Pertuzumab IgA Antibodies
2.2. Binding Affinity Studies
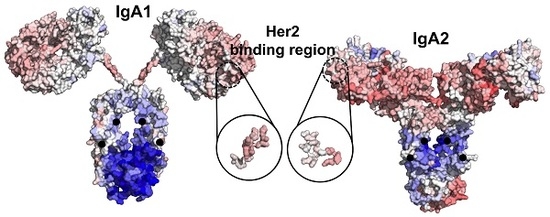
2.3. Modelling Full Antibody Structures of IgA1 and IgA2
2.4. Quantification of Allosteric Effects
2.5. Data Availability
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Strebhardt, K.; Ullrich, A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer 2008, 8, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, P. Über den jetzigen Stand der Karzinomforschung. In Beiträge zur Experimentellen Pathologie und Chemotherapie; Akademische Verlagsgesellschaft: Leipzig, Germany, 1909; pp. 117–164. [Google Scholar]
- Strohl, W.R. Current progress in innovative engineered antibodies. Protein Cell 2018, 9, 86–120. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Ishii, S.; Tachibana, T.; Maeda, A.; Higuchi, Y.; Shimaoka, S.; Moriyama, C.; Watanabe, T.; Takubo, R.; Doi, Y.; et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 2010, 28, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.; Ke, N.; Lobstein, J.; Peterson, C.; Szkodny, A.; Mansell, T.; Tuckey, C.; Riggs, P.; Colussi, P.; Noren, C.; et al. Efficient expression of full-length antibodies in the cytoplasm of engineered bacteria. Nat. Commun. 2015, 6, 8072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Yoon, H.; Jung, S. The highly evolvable Antibody Fc domain. Trends Biotechnol. 2016, 34, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Kim, J.; Kim, Y. Immunoglobulin Fc heterodimer platform technology: From design to applications in therapeutic antibodies and proteins. Front. Immunol. 2016, 7, 394. [Google Scholar] [CrossRef] [PubMed]
- Leusen, J.H. IgA as therapeutic antibody. Mol. Immunol. 2015, 68, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Valerius, T.; Stockmeyer, B.; van Spriel, A.; Graziano, R.; van den Herik-Oudijk, I.; Repp, R.; Deo, Y.; Lund, J.; Kalden, J.; Gramatzki, M.; et al. FcαRI (CD89) as a novel trigger molecule for bispecific antibody therapy. Blood 1997, 90, 4485–4492. [Google Scholar] [PubMed]
- Bakema, J.E.; Egmond, M.V. Immunoglobulin A. mAbs 2011, 3, 352–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, H.J.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woof, J.M.; Kerr, M.A. IgA function—Variations on a theme. Immunology 2004, 113, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Cerutti, A. The function and regulation of Immunoglobulin D. Curr. Opin. Immunol. 2011, 23, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Preud’homme, J.-L.; Petit, I.; Barra, A.; Morel, F.; Lecron, J.-C.; Lelievre, E. Structural and functional properties of membrane and secreted IgD. Mol. Immunol. 2000, 37, 871–887. [Google Scholar] [CrossRef]
- Bonner, A.; Perrier, C.; Corthesy, B.; Perkins, S.J. Solution structure of human secretory component and implications for biological function. J. Biol. Chem. 2007, 282, 16969–16980. [Google Scholar] [CrossRef] [PubMed]
- Bonner, A.; Furtado, P.; Almogren, A.; Kerr, M.; Perkins, S. Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy. J. Immunol. 2008, 180, 1008–1018. [Google Scholar] [CrossRef] [PubMed]
- Kelton, W.; Mehta, N.; Charab, W.; Lee, J.; Lee, C.-H.; Kojima, T.; Kang, T.H.; Georgiou, G. IgGA: A “Cross-Isotype” engineered human Fc antibody domain that displays both IgG-like and IgA-like effector functions. Chem. Biol. 2014, 21, 1603–1609. [Google Scholar] [CrossRef] [PubMed]
- Borrok, M.; Mody, N.; Lu, X.; Kuhn, M.; Wu, H.; Dall’Acqua, W.; Tsui, P. An “Fc-Silenced” IgG1 format with extended half-life designed for improved stability. J. Pharm. Sci. 2017, 106, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Igawa, T.; Maeda, A.; Haraya, K.; Tachibana, T.; Iwayanagi, Y.; Mimoto, F.; Higuchi, Y.; Ishii, S.; Tamba, S.; Hironiwa, N.; et al. Engineered monoclonal antibody with novel antigen-sweeping activity in vivo. PLoS ONE 2013, 8, e63236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Nussinov, R.; Ma, B. Antigen induced dynamic conformation changes of antibody to facilitate recognition of Fc receptors. Biophys. J. 2018, 114 (Suppl. 1), 233a. [Google Scholar] [CrossRef]
- Janda, A.; Bowen, A.; Greenspan, N.S.; Casadevall, A. Ig constant region effects on Variable region structure and function. Front. Microbiol. 2016, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.; Kozono, H.; Morii, H.; Azuma, T. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int. Immunol. 2003, 15, 417–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Kroe-Barrett, R.; Singh, S.; Roberts, C.J.; Laue, T.M. IgG cooperativity—Is there allostery? Implications for antibody functions and therapeutic antibody development. mAbs 2017, 9, 1231–1252. [Google Scholar] [CrossRef] [PubMed]
- Lua, W.; Ling, W.; Yeo, J.; Poh, J.; Lane, D.; Gan, S. The effects of antibody engineering CH and CL in Trastuzumab and Pertuzumab recombinant models: Impact on antibody production and antigen-binding. Sci. Rep. 2018, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.-L.; Lua, W.-H.; Poh, J.-J.; Yeo, J.Y.; Lane, D.P.; Gan, S.K.-E. Effect of VH-VL families in Pertuzumab and Trastuzumab recombinant production, Her2 and FcγIIA binding. Front. Immunol. 2018, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.; Appel, R.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Sim, J.Z.; Nguyen, P.V.; Lee, H.K.; Gan, S. GelApp: Mobile gel electrophoresis analyser. Nat. Methods Appl. Notes 2015. [Google Scholar] [CrossRef]
- Lua, W.H.; Gan, S.; Lane, D.; Verma, C. A search for synergy in the binding kinetics of Trastuzumab and Pertuzumab whole and F(ab) to Her2. NPJ Breast Cancer 2015, 11, 15012. [Google Scholar] [CrossRef] [PubMed]
- Rotkiewicz, P.; Skolnick, J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem. 2008, 29, 1460–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krivov, G.; Shapovalov, M.; Dunbrack, R.J. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 2009, 77, 778–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Berryman, J.T.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; et al. AMBER 14; University of California: San Francisco, CA, USA, 2015. [Google Scholar]
- Guarnera, E.; Tan, Z.W.; Zheng, Z.; Berezovsky, I.N. AlloSigMA: Allosteric Signalling and Mutation Analysis server. Bioinformatics 2017, 33, 3996–3998. [Google Scholar] [CrossRef] [PubMed]
- Chiang, R.; Gan, S.; Su, C. A computational study for rational HIV-1 non-nucleoside Reverse Transcriptase inhibitor selection and discovery of novel allosteric pockets for inhibitor design. Biosci. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.T.; Kwoh, C.K.; Verma, C.S.; Gan, S.K.E. Modeling the full length HIV-1 Gag polyprotein reveals the role of its p6 subunit in viral maturation and the effect of non-cleavage site mutations in protease drug resistance. J. Biomol. Struct. Dyn. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kurochkin, I.V.; Guarnera, E.; Wong, J.H.; Eisenhaber, F.; Berezovsky, I.N. Toward allosterically increased catalytic activity of insulin-degrading enzyme against amyloid peptides. Biochemistry 2017, 56, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Guarnera, E.; Berezovsky, I.N. Structure-based statistical mechanical model accounts for the causality and energetics of allosteric communication. PLoS Comput. Biol. 2016, 12, e1004678. [Google Scholar] [CrossRef] [PubMed]
- Feige, M.J.; Hendershot, L.M.; Buchner, J. How antibodies fold. Trends Biochem. Sci. 2009, 35, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Hopf, T.A.; Colwell, L.J.; Sheridan, R.; Rost, B.; Sander, C.; Marks, D.S. Three-dimensional structures of membrane proteins from genomic sequencing. Cell 2012, 149, 1607–1621. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Dunbar, J.; Alcala, M.; Deane, C.M. Variable regions of antibodies and T-cell Receptors may not be sufficient in molecular simulations investigating binding. J. Chem. Theory Comput. 2017, 13, 3097–3105. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Oakman, E.L.; Thorpe, I.F.; Shi, X.; Abbyad, C.L.B.; Boxer, S.G.; Romesberg, F.E. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. USA 2006, 103, 13722–13727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, L.T.; Duan, Y.; Wang, L.; Massova, I.; Kollman, P.A. Molecular dynamics and free-energy calculations applied to affinity maturation in antibody 48G7. Proc. Natl. Acad. Sci. USA 1999, 96, 14330–14335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Tracka, M.B.; Uddin, S.; Casas-Finet, J.; Jacobs, D.J.; Livesay, D.R. Rigidity emerges during Antibody evolution in three distinct antibody systems: Evidence from QSFR analysis of Fab fragments. PLoS Comput. Biol. 2015, 11, e1004327. [Google Scholar] [CrossRef] [PubMed]
- Midelfort, K.; Hernandez, H.; Lippow, S.; Tidor, B.; Drennan, C.; Wittrup, K. Substantial energetic improvement with minimal structural perturbation in a high affinity mutant antibody. J. Mol. Biol. 2004, 343, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Jeliazkov, J.R.; Sljoka, A.; Kuroda, D.; Tsuchimura, N.; Katoh, N.; Tsumoto, K.; Gray, J.J. Repertoire analysis of Antibody CDR-H3 loops suggests affinity maturation does not typically result in rigidfication. Front. Immunol. 2018, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Su, C.T.-T.; Ling, W.-L.; Lua, W.-H.; Poh, J.-J.; Gan, S.K.-E. The role of antibody Vκ Framework 3 region towards antigen binding: Effects on recombinant production and Protein L binding. Sci. Rep. 2017, 7, 3766. [Google Scholar] [CrossRef] [PubMed]
- Phua, S.-X.; Lua, W.-H.; Gan, S.K.-E. Role of FcaR EC2 region in extracellular membrane localization. Cell Cycle 2018. [Google Scholar] [CrossRef] [PubMed]


| Δgregion (kcal/mol) | ||||
|---|---|---|---|---|
| Mutating Event | Her2-Binding Event | |||
| Region | IgA1 | IgA2 | IgA1 | IgA2 |
| Her2 binding site | 0.18 | 0.12 | 0.06 | −0.05 |
| Fab | 0.15 | 0.38 | 0.08 | 0.02 |
| Fc (CH2–CH3) | −0.46 | −0.08 | 0.03 | −0.007 |
| Hinge | 0.05 | 0.04 | −0.02 | 0.03 |
| All Heavy Atoms/Cα Atoms | ||
|---|---|---|
| IgA1 | IgA2 | |
| Wild type | 100/100 | 100/100 |
| Mutant (replicate 1) | 69.7/58.3 | 57.1/61.8 |
| Mutant (replicate 2) | 66.5/56.7 | 55.7/61.8 |
| Mutant (replicate 3) | 66.5/56.7 | 54.7/61.8 |
| Δgregion (kcal/mol) in the Event of Mutations | ||||||
|---|---|---|---|---|---|---|
| IgA1 (C266/H317) | IgA2 (C253/H304) | IgA1 (C266) | IgA2 (C253) | IgA1 (H317) | IgA2 (H304) | |
| Her2 site | 0.38 ± 0.32 | 0.12 ± 0.06 | 0.18 ± 0.16 | 0.06 ± 0.07 | 0.28 ± 0.23 | 0.09 ± 0.05 |
| Fab | 0.62 ± 0.54 | 0.31 ± 0.12 | 0.28 ± 0.26 | 0.23 ± 0.09 | 0.5 ± 0.36 | 0.13 ± 0.08 |
| Fc (CH2–CH3) | −0.52 ± 0.05 | −0.16 ± 0.27 | −0.4 ± 0.07 | −0.06 ± 0.29 | −0.25 ± 0.09 | −0.1 ± 0.03 |
| Hinge | 0.21 ± 0.2 | 0.08 ± 0.05 | 0.08 ± 0.09 | −0.003 ± 0.032 | 0.20 ± 0.17 | 0.07 ± 0.04 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, C.T.-T.; Lua, W.-H.; Ling, W.-L.; Gan, S.K.-E. Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding. Antibodies 2018, 7, 20. https://doi.org/10.3390/antib7020020
Su CT-T, Lua W-H, Ling W-L, Gan SK-E. Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding. Antibodies. 2018; 7(2):20. https://doi.org/10.3390/antib7020020
Chicago/Turabian StyleSu, Chinh Tran-To, Wai-Heng Lua, Wei-Li Ling, and Samuel Ken-En Gan. 2018. "Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding" Antibodies 7, no. 2: 20. https://doi.org/10.3390/antib7020020
APA StyleSu, C. T. -T., Lua, W. -H., Ling, W. -L., & Gan, S. K. -E. (2018). Allosteric Effects between the Antibody Constant and Variable Regions: A Study of IgA Fc Mutations on Antigen Binding. Antibodies, 7(2), 20. https://doi.org/10.3390/antib7020020