Dried Blood Spot in Laboratory: Directions and Prospects
Abstract
:1. Introduction
2. Search Strategy
3. Types of Membrane Carriers, Biomaterial Collection, Processing, Storage and Logistics
4. Modern Analytical Methods for Dried Blood Spot Analysis
4.1. RNA/DNA Quantification
4.2. Protein/Peptide Detection
4.3. Metabolite Detection
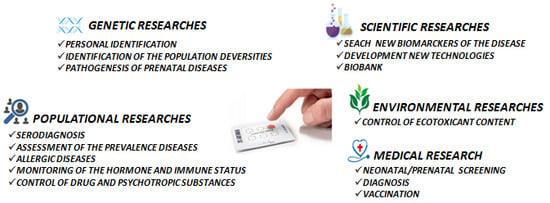
5. Practical Applications
5.1. Newborn Screening
5.2. Serodiagnosis
5.3. Bioenvironmental Monitoring
6. Limitation of DBS in Biomedical Research
7. Biobank
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gruner, N.; Stamboul, O.; Ross, S.R. Dried Blood Spots—Preparing and Processing for Use in Immunoassays and in Molecular Techniques. J. Vis. Exp. 2015, 97, 52619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisitsa, A.V.; Ponomarenko, E.A.; Lokhov, P.G.; Archakov, A.I. Postgenomic Medicine: Alternative to Biomarkers. Ann. Russ. Acad. Med. Sci. 2016, 71, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, A.; Rosenstein, M.; Kondakov, S.; Cherevko, N. Diagnosis of food hypersensitivity mediated through тyре III immunopathological reactions. Russ. J. Immunol. 2015, 9, 150–153. [Google Scholar]
- Kopylov, A.; Ponomarenko, E.; Ilgisonis, E.; Pyatnitskiy, M.; Lisitsa, A.; Poverennaya, E.; Kiseleva, O.; Farafonova, T.; Tikhonova, O.; Zavialova, M.; et al. 200+ Protein Concentrations in Healthy Human Blood Plasma: Targeted Quantitative SRM SIS Screening of Chromosomes 18, 13, Y and the Mitochondrial Chromosome Encoded Proteome. J. Proteome Res. 2018, 18, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Larina, I.; Pastushkova, L.; Kireev, K.; Grigoriev, A. The formation of the urine proteome in healthy human (review). Fiziologiya Cheloveka 2013, 39, 43–59. [Google Scholar]
- Tu, C.; Rudnick, P.A.; Martinez, M.Y.; Cheek, K.L.; Stein, S.E.; Slebos, R.J.; Liebler, D.C. Depletion of Abundant Plasma Proteins and Limitations of Plasma. J. Proteome Res. 2010, 9, 4982–4991. [Google Scholar] [CrossRef] [Green Version]
- Alonso, S.G.; de la Torre Díez, I.; Zapiraín, B.G. Predictive, Personalized, Preventive and Participatory (4P) Medicine Applied to Telemedicine and eHealth in the Literature. J. Med. Syst. 2019, 43, 140. [Google Scholar] [CrossRef]
- Horgan, R.; Kenny, L. ‘Omic’ technologies: Genomics, transcriptomics, proteomics and metabolomics. TOG 2011, 13, 189–195. [Google Scholar] [CrossRef]
- Geyer Ph Holdt, L.; Teupser, D.; Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13, 942. [Google Scholar] [CrossRef]
- Demirev, P. Dried blood spots: Analysis and applications. J. Anal. Chem. 2013, 85, 779–789. [Google Scholar] [CrossRef]
- Meesters, R.; Hooff, G. State-of-the-art dried blood spot analysis: An overview of recent advances and future trends. Bioanalysis 2013, 5, 2187–2208. [Google Scholar] [CrossRef]
- Konig, S.; Yildiz, O.; Hermann, N.; Steurer, A.; Singrasa, M.; Dobelin, W. A Novel Concept for Sample Collection and Sample Preparation; Poster WP 427; ASMS: Salt Lake City, UT, USA, 2010. [Google Scholar]
- Rottinghaus, E.; Beard, S.; Bile, E.; Modukanele, M.; Maruping, M.; Mine, M.; Nkengasong, J.; Yang, C. Evaluation of Dried Blood Spots Collected on Filter Papers from Three Manufacturers Stored at Ambient Temperature for Application in HIV-1 Drug Resistance Monitoring. PLoS ONE 2014, 9, e109060. [Google Scholar] [CrossRef] [PubMed]
- Mwaba, P.; Cassol, S.; Nunn, A.; Pilon, R.; Chintu, C.; Janes, M.; Zumla, A. Whole blood versus plasma spots for measurement of HIV-1 viral load in HIV-infected African patients. Lancet 2003, 362, 2067–2068. [Google Scholar] [CrossRef]
- Wagner, M.; Tonoli, D.; Varesio, E.; Hopfgartner, G. The use of mass spectrometry to analyze dried blood spots. Mass Spectrom. Rev. 2014, 35, 361–438. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Lejczak, C.; Lambert, C.; Servais, J.; Makombe, N.; Rusine, J.; Staub, T.; Hemmer, R.; Schneider, F.; Schmit, J.; et al. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J. Clin. Microbiol. 2004, 42, 16–20. [Google Scholar] [CrossRef] [Green Version]
- Mwaba, P. Dried blood spot technology for CD4+ T-cell counting. Lancet 2004, 363, 1074–1075. [Google Scholar] [CrossRef]
- Brambilla, D.; Jennings, C.; Aldrovandi, G.; Bremer, J.; Comeau, A.; Cassol, S.; Dickover, R.; Jackson, J.; Pitt, J.; Sullivan, J.; et al. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: Measurement, precision, and RNA stability. J. Clin. Microbiol. 2003, 41, 1888–1893. [Google Scholar] [CrossRef] [Green Version]
- Cassol, S.; Read, S.; Weniger, B.; Gomez, P.; Lapointe, N.; Ou, C.; Babu, P. Dried blood spots collected on filter paper: An international resource for the diagnosis and genetic characterization of human immunodeficiency virus type-1. Memórias do Instituto Oswaldo Cruz 1996, 91, 351–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, N.; Mondal, T.; Preissler, M.; Freed, B.; Stockinger, S.; Bell, E.; Druschel, C.; Louis, G.; Lawrence, D. Detection of immunoglobulin isotypes from dried blood spots. J. Immunol. Methods 2014, 404, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Kivuyo, S.L.; Johannessen, A.; Trøseid, M.; Kasubi, M.J.; Gundersen, S.G.; Naman, E.; Mushi, D.; Ngowi, B.J.; Mfinanga, G.S.; Bruun, J.N. p24 antigen detection on dried blood spots is a feasible and reliable test for infant HIV infection in rural Tanzania. Int. J. STD AIDS 2011, 22, 719–721. [Google Scholar] [CrossRef]
- Deursen, P.; Oosterlaken, T.; Andre, P.; Verhoeven, A.; Bertens, L.; Trabaud, M.; Ligeon, V.; Jong, J. Measuring human immunodeficiency virus type 1 RNA loads in dried blood spot specimens using NucliSENS EasyQ HIV-1 v2.0. J. Clin. Virol. 2010, 47, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Rottinghaus, E.; Bile, E.; Modukanele, M.; Maruping, M.; Mine, M.; Nkengasong, J.; Yang, C. Comparison of Ahlstrom Grade 226, Munktell TFN, and Whatman 903 Filter Papers for Dried Blood Spot Specimen Collection and Subsequent HIV-1 Load and Drug Resistance Genotyping Analysis. ASM 2013, 51, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Beaudette, P.; Bateman, K. Discovery stage pharmacokinetics using dried blood spots. J. Chrom. B J. Life Sci. 2004, 809, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Koal, T.; Burhenne, H.; Romling, R.; Svoboda, M.; Resch, K.; Kaever, V. Quantification of antiretroviral drugs in dried blood spot samples by means of liquid chromatography/tandem mass spectrometry. RCM 2005, 19, 2995–3001. [Google Scholar] [CrossRef] [PubMed]
- Behets, F.; Kashamuka, M.; Pappaioanou, M.; Green, T.; Ryder, R.; Batter, V.; George, J.; Hannon, W.; Quinn, T. Stability of human immunodeficiency virus type 1 antibodies in whole blood dried on filter paper and stored under various tropical conditions in Kinshasa, Zaire. J. Clin. Microbiol. 1992, 30, 1179–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evengard, B.; Ehrnst, A.; von Sydow, M.; Pehrson, P.; Lundbergh, P.; Linder, E. Effect of heat on extracted HIV viral infectivity and antibody activity using the filter paper technique of blood sampling. AIDS 1989, 3, 591–595. [Google Scholar] [CrossRef]
- Cassol, S.; Gill, M.; Pilon, R.; Cormier, M.; Voigt, R.; Willoughby, B.; Forbes, J. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J. Clin. Microbiol. 1997, 35, 2795–2801. [Google Scholar] [CrossRef] [Green Version]
- Chao, T.; Arjmandi-Tash, O.; Das, D.; Starov, V. Spreading of blood drops over dry porous substrate: Complete wetting case. J. Colloid Interface Sci. 2015, 446, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, S.; Delaby, C.; Vialaret, J.; Ducos, J.; Hirtz, C. Current and future use of “dried blood spot” analyses in clinical chemistry. CCLM 2013, 51, 1897–1909. [Google Scholar] [CrossRef]
- Taniguchi, M.; Pieracci, J.; Belfort, G. Effect of Undulations on Surface Energy: Quantitative Assessment. Langmuir 2001, 17, 4312–4315. [Google Scholar] [CrossRef]
- Samsonov, J.; Senatova, S.; Muratov, D.; Osipov, A.; Kondakov, S.; Kuznetsov, D. Modification of the membrane materials used in the technology of dry blood stains, zinc oxide nanoparticles. Mosc. Univ. Bull. 2015, 56, 418–423. [Google Scholar]
- Adam, B.W.; Alexander, J.R.; Smith, S.J.; Chace, D.H.; Loeber, J.G.; Elvers, L.H.; Hannon, W.H. Recoveries of phenylalanine from two sets of dried-blood-spot reference materials: Prediction from hematocrit, spot volume, and paper matrix. Clin. Chem. 2000, 46, 126–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youhnovski, N.; Bergeron, A.; Furtado, M.; Garofolo, F. Pre-cut dried blood spot (PCDBS): An alternative to dried blood spot (DBS) technique to overcome hematocrit impact. Rapid Commun. Mass Spectrom. 2011, 25, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Paehler, T.; Zimmer, M.; Guo, Z.; Zane, P.; Emmons, G.T. Impact of various factors on radioactivity distribution in different DBS papers. Bioanalysis 2010, 2, 1469–1475. [Google Scholar] [CrossRef]
- Denniff, P.; Spooner, N. The effect of hematocrit on assay bias when using DBS samples for the quantitative bioanalysis of drugs. Bioanalysis 2010, 2, 1385–1395. [Google Scholar] [CrossRef]
- Timmerman, P.; White, S.; Cobb, Z.; Woods, K.; de Vries, R.; Spooner, N.; Sangster, T.; Dillen, L.; Hawthorne, G. European Bioanalysis Forum continued plans to support liquid microsampling. Bioanalysis 2014, 6, 1897–1900. [Google Scholar] [CrossRef]
- Hall, E.M.; Flores, S.R.; De Jesús, V.R. Influence of Hematocrit and Total-Spot Volume on Performance Characteristics of Dried Blood Spots for Newborn Screening. Int. J. Neonatal Screen. 2015, 1, 69–78. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. NBS01-A6 Blood Collection on Filter Paper for Newborn Screening Programs, 6th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013. [Google Scholar]
- Moretti, M.; Freni, F.; Tomaciello, I.; Vignali, C.; Groppi, A.; Visonà, S.D.; Tajana, L.; Osculati, A.M.M.; Morini, L. Determination of benzodiazepines in blood and in dried blood spots collected from post-mortem samples and evaluation of the stability over a three-month period. Drug Test. Anal. 2019, 11, 1403–1411. [Google Scholar] [CrossRef]
- Moretti, M.; Visonà, S.D.; Freni, F.; Tomaciello, I.; Vignali, C.; Groppi, A.; Tajana, L.; Osculati, A.M.M.; Morini, L. A liquid chromatography-tandem mass spectrometry method for the determination of cocaine and metabolites in blood and in dried blood spots collected from postmortem samples and evaluation of the stability over a 3-month period. Drug Test. Anal. 2018, 10, 1430–1437. [Google Scholar] [CrossRef]
- Moretti, M.; Freni, F.; Valentini, B.; Vignali, C.; Groppi, A.; Visonà, S.D.; Osculati, A.M.M.; Morini, L. Determination of Antidepressants and Antipsychotics in Dried Blood Spots (DBSs) Collected from Post-Mortem Samples and Evaluation of the Stability over a Three-Month Period. Molecules 2019, 24, 3636. [Google Scholar] [CrossRef] [Green Version]
- Irie, K.; Shobu, S.; Hiratsuji, S.; Yamasaki, Y.; Nanjo, S.; Kokan, C.; Hata, A.; Kaji, R.; Masago, K.; Fujita, S.; et al. Development and validation of a method for gefitinib quantification in dried blood spots using liquid chromatography-tandem mass spectrometry: Application to finger-prick clinical blood samples of patients with non-small cell lung cancer. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1087–1088, 1–5. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, H.; Huang, X.; Shao, L.; Xie, X.; Wang, F.; Yang, J.; Pei, P.; Zhang, Z.; Zhai, Y.; et al. A novel LC-MS/MS assay for vitamin B1, B2 and B6 determination in dried blood spots and its application in children. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1112, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gohring, K.; Dietz, K.; Hartleif, S.; Jahn, G.; Hamprecht, K. Influence of different extraction methods and PCR techniques on the sensitivity of HCMV-DNA detection in dried blood spot (DBS) filter cards. J. Clin. Virol. 2010, 48, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Strenger, V.; Pfurtscheller, K.; Wendelin, G.; Aberle, S.W.; Nacheva, E.P.; Zöhrer, B.; Zenz, W.; Nagel, B.; Zobel, G.; Popow-Kraupp, T. Differentiating inherited human herpesvirus type 6 genome from primary human herpesvirus type 6 infection by means of dried blood spot from the newborn screening card. IJP 2011, 159, 859–861. [Google Scholar] [CrossRef]
- Villar, L.M.; Cruz, H.M.; Deodato, R.M.; Miguel, J.C.; da Silva, E.F.; Flores, G.L.; Lewis-Ximenez, L.L. Usefulness of automated assays for detecting hepatitis B and C markers in dried blood spot samples. BMC Res. Notes 2019, 12, 523. [Google Scholar] [CrossRef]
- Kenmoe, S.; Tagnouokam PA, N.; Nde, C.K.; Mella-Tamko, G.F.; Njouom, R. Using dried blood spot for the detection of HBsAg and anti-HCV antibodies in Cameroon. BMC Res. Notes 2018, 11, 818. [Google Scholar] [CrossRef] [PubMed]
- Lakshmy, R.; Gupta, R. Measurement of glycated hemoglobin A1c from dried blood by turbidimetric immunoassay. JDST 2009, 3, 1203–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fachiroh, J.; Prasetyanti, P.R.; Paramita, D.K.; Prasetyawati, A.T.; Anggrahini, D.W.; Haryana, S.M.; Middeldorp, J.M. Dried-blood sampling for epstein-barr virus immunoglobulin G (IgG) and IgA serology in nasopharyngeal carcinoma screening. J. Clin. Microbiol. 2008, 46, 1374–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helfand, R.; Cabezas, C.; Abernathy, E.; Castillo-Solorzano, C.; Ortiz, A.; Sun, H.; Osores, F.; Oliveira, L.; Whittembury, A.; Charles, M.; et al. Dried Blood Spots versus Sera for Detection of Rubella Virus-Specific Immunoglobulin M (IgM) and IgG in Samples Collected during a Rubella Outbreak in Peru. CVI 2007, 14, 1522–1525. [Google Scholar] [CrossRef] [Green Version]
- Balmaseda, A.; Saborio, S.; Tellez, Y.; Mercado, J.C.; Pérez, L.; Hammond, S.N.; Rocha, C.; Kuan, G.; Harris, E. Evaluation of immunological markers in serum, filter-paper blood spots, and saliva for dengue diagnosis and epidemiological studies. J. Clin. Virol. 2008, 43, 287–291. [Google Scholar] [CrossRef]
- Tuaillon, E.; Mondain, A.M.; Meroueh, F.; Ottomani, L.; Picot, M.C.; Nagot, N.; Van de Perre, P.; Ducos, J. Dried blood spot for hepatitis C virus serology and molecular testing. Hepatol 2010, 51, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Moron, S.; Ryan, P.; Ardizone-Jiménez, B.; Martín, D.; Troya, J.; Cuevas, G.; Valencia, J.; Jimenez-Sousa, M.; Avellón, A.; Resino, S. Evaluation of dried blood spot samples for screening of hepatitis C and human immunodeficiency virus in a real-world setting. Sci. Rep. 2018, 8, 1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, J.; Becker, C.; Persson, N.; Fex, M.; Torn, C. C-peptide in dried blood spots. Scand. J. Clin. Lab. Investig. 2010, 70, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Kroll Ch Ferber, M.; Dawson, B.; Jacobson, R.; Mensink, K.; Lorey, F.; Sherwin, J.; Cunningham, G.; Rinaldo, P.; Matern, D.; Hahn, S. Retrospective determination of ceruloplasmin in newborn screening blood spots of patients with Wilson disease. Mol. Genet. Metab. 2006, 89, 134–186. [Google Scholar] [CrossRef]
- Chambers, A.; Percy, A.; Yang, J.; Camenzind, A.; Borchers, C. Multiplexed quantitation of endogenous proteins in dried blood spots by multiple reaction monitoring mass spectrometry. Mol. Cell Proteom. 2012, 12, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, A.; Butcher, P.; Maden, K.; Walker, S.; Widmer, M. Semi-targeted residue screening in complex matrices with liquid chromatography coupled to high resolution mass spectrometry: Current possibilities and limitations. Analyst 2011, 136, 1898–1909. [Google Scholar] [CrossRef]
- de Sain-van der Velden, M.G.M.; Ham, M.; Gerrits, J.; Prinsen, H.; Willemsen, M.; Pras-Raves, M.; Jans, J.; Verhoeven-Duif, N. Quantification of metabolites in dried blood spots by direct infusion high resolution mass spectrometry. Anal. Chim. Act. 2017, 979, 45–50. [Google Scholar] [CrossRef]
- Coelho, M.; Mendes, V.; Limab, I.; Martinsa, F.; Fernandesb, A.; Macedob, M.; Jonesa, J.; Manadas, B. Direct analysis of [6,6-2H2]glucose and [U-13C6]glucose dry blood spotenrichments by LC–MS/MS. J. Chrom. B 2016, 1022, 242–248. [Google Scholar] [CrossRef]
- Zimmermann, M.; Morreti, D.; Chaoki, N.; Torresani, T. Development of a Dried Whole- Blood Spot Thyroglobulin Assay and Its Evaluation as an Indicator of Thyroid Status in Goitrous Children Receiving Iodized Salt. AJCN 2003, 77, 1453–1458. [Google Scholar] [CrossRef] [Green Version]
- Skogstrand, K.; Thornsen, P.; Norgaard-Pedersen, B.; Schendel, D.; Sorensen, L.; Hougaard, D. Simultaneous Measurement of 25 Infl ammatory Markers and Neurotrophins in Neonatal Dried Blood Spots by Immunoassay With xMAP Technology. Clin. Chem. 2005, 51, 1854–1866. [Google Scholar] [CrossRef]
- Odoardi, S.; Anzillotti, L.; Strano-Rossi, S. Simplifying sample pretreatment: Application of dried blood spot (DBS) method to blood samples, including postmortem, for UHPLC–MS/MS analysis of drugs of abuse. Forensic Sci. Int. 2014, 243, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Simões, S.; Ajenjo, A.; Dias, M.J. Dried blood spots combined to an UPLC-MS/MS method for the simultaneous determination of drugs of abuse in forensic toxicology. J. Pharm. Biomed. Anal. 2018, 147, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, R.; Allen, K.J.; Koplin, J.J.; Roche, P.; Greaves, R.F. Advantages and Challenges of Dried Blood Spot Analysis by Mass Spectrometry Across the Total Testing Process. EJIFCC 2016, 27, 288–317. [Google Scholar] [PubMed]
- Heinig, K.; Bucheli, F.; Hartenbach, R.; Gajate-Perez, A. Determination of mycophenolic acid and its phenyl glucuronide in human plasma, ultrafiltrate, blood, DBS and dried plasma spots. Bioanalysis 2010, 2, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tse, F.L. Dried blood spot sampling in combination with LC-MS/MS for quantitative analysis of small molecules. Biomed. Chromatogr. 2010, 24, 49–65. [Google Scholar] [CrossRef] [PubMed]
- McDade, T.; Williams, S.; Snodgrass, J. What a drop can do: Dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 2007, 44, 899–925. [Google Scholar] [CrossRef] [Green Version]
- Caggana, M.; Jones, E.A.; Shahied, S.I.; Tanksley, S.; Hermerath, C.A.; Lubin, I.M. Newborn screening: From Guthrie to whole genome sequencing. Public Health Rep. 2013, 128, 14–19. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC). Ten great public health achievements—United States, 2001–2010. MMWR Morb. Mortal. Wkly. Rep. 2011, 60, 619–623. [Google Scholar]
- Mak, C.M.; Lee, H.C.; Chan, A.Y.; Lam, C.W. Inborn errors of metabolism and expanded newborn screening: Review and update. Crit. Rev. Clin. Lab. Sci. 2013, 50, 142–162. [Google Scholar] [CrossRef]
- The Clinical and Laboratory Standards Institute. Newborn Screening by Tandem Mass Spectrometry; Approved Guideline; The Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Bodamer, O.A. Screening for Phenylketonuria. Ann. Nestlé 2010, 68, 53–57. [Google Scholar] [CrossRef]
- Gelb, M.H.; Lukacs, Z.; Ranieri, E.; Schielen, P.C.J.I. Newborn Screening for Lysosomal Storage Disorders: Methodologies for Measurement of Enzymatic Activities in Dried Blood Spots. Int. J. Neonatal Screen. 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Céspedes, N.; Valencia, A.; Echeverry, C.A.; Arce-Plata, M.I.; Colón, C.; Castiñeiras, D.E.; Hurtado, P.M.; Cocho, J.A.; Herrera, S.; Arévalo-Herrera, M. Reference values of amino acids, acylcarnitines and succinylacetone by tandem mass spectrometry for use in newborn screening in southwest Colombia. Colomb. Médica 2017, 48, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Kirk, E.P.; Fletcher, J.M.; Sharp, P.; Carey, B.; Poulos, A. X-linked adrenoleukodystrophy: The Australasian experience. Am. J. Med. Genet. 1998, 76, 420–423. [Google Scholar] [CrossRef]
- Bezman, L.; Moser, A.B.; Raymond, G.V.; Rinaldo, P.; Watkins, P.A.; Smith, K.D.; Kass, N.E.; Moser, H.W. Adrenoleukodystrophy: Incidence, new mutation rate, and results of extended family screening. Ann. Neurol. 2001, 49, 512–517. [Google Scholar] [CrossRef]
- Kemper, A.R.; Brosco, J.; Comeau, A.M.; Green, N.S.; Grosse, S.D.; Jones, E.; Kwon, J.M.; Lam, W.K.; Ojodu, J.; Prosser, L.A.; et al. Newborn screening for X-linked adrenoleukodystrophy: Evidence summary and advisory committee recommendation. Genet. Med. 2017, 19, 121–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard, C.; Gaspar, H.B.; Al-Herz, W.; Bousfiha, A.; Casanova, J.L.; Chatila, T.; Crow, Y.J.; Cunningham-Rundles, C.; Etzioni, A.; Franco, J.L.; et al. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 2018, 38, 96–128. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.; Puck, J.M. Development of population-based newborn screening for severe combined immunodeficiency. J. Allergy Clin. Immunol. 2005, 115, 391–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Burg, M.; Tümkaya, T.; Boerma, M.; de Bruin-Versteeg, S.; Langerak, A.W.; van dongen, J.J. Ordered recombination of immunoglobulin light chain genes occurs at the IGK locus but seems less strict at the IGL locus. Blood 2001, 97, 1001–1008. [Google Scholar] [CrossRef]
- van Zelm, M.C.; Szczepanski, T.; van der Burg, M.; van Dongen, J.J. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J. Exp. Med. 2007, 204, 645–655. [Google Scholar] [CrossRef]
- van der Burg, M.; Mahlaoui, N.; Gaspar, H.B.; Pai, S.Y. Universal Newborn Screening for Severe Combined Immunodeficiency (SCID). Front. Pediatr. 2019, 7, 373. [Google Scholar] [CrossRef]
- Barbaro, M.; Ohlsson, A.; Borte, S.; Jonsson, S.; Zetterström, R.H.; King, J.; Winiarski, J.; von Döbeln, U.; Hammarström, L. Newborn Screening for Severe Primary Immunodeficiency Diseases in Sweden-a 2-Year Pilot TREC and KREC Screening Study. J. Clin. Immunol. 2017, 37, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Czibere, L.; Burggraf, S.; Fleige, T.; Glück, B.; Keitel, L.M. High-throughput genetic newborn screening for spinal muscular atrophy by rapid nucleic acid extraction from dried blood spots and 384- well qPCR. Eur. J. Hum. Genet. 2020, 28, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.; Stambouli, O.; Grüner, N.; Marcus, U.; Cai, W.; Zhang, W.; Zimmermann, R.; Roggendorf, M. Detection of infections with hepatitis B virus, hepatitis C virus and human immunodeficiency virus by analyses of dried blood spots—Performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol. J. 2013, 10, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, S.; Cubitt, W. The use of the dried blood spot sample in epidemiological studies. J. Clin. Pathol. 1999, 52, 633–639. [Google Scholar] [CrossRef] [Green Version]
- Hofman, L.; Foley, T.; Henry, J.; Naylor, E. Assays for thyroid-stimulating hormone using dried blood spotted filter paper specimens to screen for hypothyroidism in older children and adults. J. Med. Screen. 2003, 10, 5–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, T.; Shackleton, C.; Covey, T.; Ellis, G. Identification of the steroids in neonatal plasma that interfere with 17 alpha-hydroxyprogesterone radioimmunoassays. Clin. Сhem. 1992, 38, 1830–1837. [Google Scholar] [CrossRef]
- Breivika, K.; Vestrengb, V.; Rozovskayac, O.; Pacynaad, J.M. Atmospheric emissions of some POPs in Europe: A discussion of existing inventories and data needs. Environ. Sci. Policy 2006, 9, 663–674. [Google Scholar] [CrossRef]
- Jaraczewska, K.; Lulek, J.; Covaci, A.; Voorspoels, S.; Kaluba-Skotarczak, A.; Drews, K.; Schepens, P. Distribution of polychlorinated biphenyls, organochlorine pesticides and polybrominated diphenyl ethers in human umbilical cord serum, maternal serum and milk from Wielkopolska region, Poland. Sci. Total Environ. 2006, 372, 20–31. [Google Scholar] [CrossRef]
- Morales, E.; Gascon, M.; Martinez, D.; Casas, M.; Ballester, F.; Rodríguez-Bernal, C.L.; Ibarluzea, J.; Marina, L.S.; Espada, M.; Goñi, F.; et al. Associations between blood persistent organic pollutants and 25-hydroxyvitamin D3 in pregnancy. Environ. Int. 2013, 57–58, 34–41. [Google Scholar] [CrossRef]
- Ha, M.H.; Lee, D.H.; Jacobs, D.R. Association between serum concentrations of persistent organic pollutants and self-reported cardiovascular disease prevalence: Results from the National Health and Nutrition Examination Survey, 1999–2002. Environ. Health Perspect. 2007, 115, 1204–1209. [Google Scholar] [CrossRef]
- Tsuji, M.; Aiko, Y.; Kawamoto, T.; Hachisuga, T.; Kooriyama, C.; Myoga, M.; Tomonaga, C.; Matsumura, F.; Anan, A.; Tanaka, M.; et al. Polychlorinated biphenyls (PCBs) decrease the placental syncytiotrophoblast volume and increase Placental Growth Factor (PlGF) in the placenta of normal pregnancy. Placenta 2013, 34, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Bertrand, K.A.; Choi, A.L.; Hu, F.B.; Laden, F.; Grandjean, P.; Sun, Q. Persistent organic pollutants and type 2 diabetes: A prospective analysis in the nurses’ health study and meta-analysis. Environ. Health Perspect. 2013, 121, 153–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valvi, D.; Mendez, M.A.; Martinez, D.; Grimalt, J.O.; Torrent, M.; Sunyer, J.; Vrijheid, M. Prenatal concentrations of polychlorinated biphenyls, DDE, and DDT and overweight in children: A prospective birth cohort study. Environ. Health Perspect. 2012, 120, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Lignell, S.; Aune, M.; Darnerud, P.O.; Hanberg, A.; Larsson, S.C.; Glynn, A. Prenatal exposure to polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs) may influence birth weight among infants in a Swedish cohort with background exposure: A cross-sectional study. Environ. Health 2013, 12, 44. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.L.; Gao, C.; Bell, E.M.; Druschel, C.M.; Caggana, M.; Aldous, K.M.; Buck Louis, G.M.; Kannan, K. Analysis of polychlorinated biphenyls and organochlorine pesticides in archived dried blood spots and its application to track temporal trends of environmental chemicals in newborns. Environ. Res. 2014, 133, 204–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.L.; Yun, S.; Bell, E.M.; Druschel, C.M.; Caggana, M.; Aldous, K.M.; Buck Louis, G.M.; Kannan, K. Temporal trends of polybrominated diphenyl ethers (PBDEs) in the blood of newborns from New York State during 1997 through 2011: Analysis of dried blood spots from the newborn screening program. Environ. Sci. Technol. 2013, 16, 47, 8015–8021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghassabian, A.; Bell, E.M.; Ma, W.L.; Sundaram, R.; Kannan, K.; Buck Louis, G.M.; Yeung, E. Concentrations of perfluoroalkyl substances and bisphenol A in newborn dried blood spots and the association with child behavior. Environ. Pollut. 2018, 243, 1629–1636. [Google Scholar] [CrossRef]
- Mohammed, B.; Cameron, G.; Cameron, L.; Hawksworth, G.; Helms, P.; McLay, J. Can-prick sampling replace venous sampling to determine the pharmacokinetic profile of oral paracetamol? Br. J. Clin. Pharmacol. 2010, 70, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Strnadova, K.; Holub, M.; Muhl, A.; Heinze, G.; Ratschmann, R.; Mascher, H.; Stockler-Ipsiroglu, S.; Waldhauser, F.; Votava, F.; Lebl, J.; et al. Long-term stability of amino acids and acylcarnitines in dried blood spots. Clin. Chem. 2007, 53, 717–722. [Google Scholar] [CrossRef] [Green Version]
- Kong, S.; Lin, H.; Ching, J.; Ho, P. Evaluation of dried blood spots as sample matrix for gas chromatography/mass spectrometry based metabolomic profiling. Anal. Chem. 2011, 83, 4314–4318. [Google Scholar] [CrossRef]
- Gutor, S.; Engleski, N.; Prokudina, D.; Oskin, S.; Gerasimov, E.; Sazonov, A.; Ogorodova, L. Bank of biosamples: Management and physical implementation. Phys. IT 2013, 4, 31–39. [Google Scholar]
- Reznik, O.; Kuzmin, D.; Skvortsov, A.; Reznik, A. Biobanks are an essential tool for transplantation. History, current state, perspectives. Vestn. Transpl. Artif. Organs. 2016, 18, 123–132. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malsagova, K.; Kopylov, A.; Stepanov, A.; Butkova, T.; Izotov, A.; Kaysheva, A. Dried Blood Spot in Laboratory: Directions and Prospects. Diagnostics 2020, 10, 248. https://doi.org/10.3390/diagnostics10040248
Malsagova K, Kopylov A, Stepanov A, Butkova T, Izotov A, Kaysheva A. Dried Blood Spot in Laboratory: Directions and Prospects. Diagnostics. 2020; 10(4):248. https://doi.org/10.3390/diagnostics10040248
Chicago/Turabian StyleMalsagova, Kristina, Artur Kopylov, Alexander Stepanov, Tatyana Butkova, Alexander Izotov, and Anna Kaysheva. 2020. "Dried Blood Spot in Laboratory: Directions and Prospects" Diagnostics 10, no. 4: 248. https://doi.org/10.3390/diagnostics10040248
APA StyleMalsagova, K., Kopylov, A., Stepanov, A., Butkova, T., Izotov, A., & Kaysheva, A. (2020). Dried Blood Spot in Laboratory: Directions and Prospects. Diagnostics, 10(4), 248. https://doi.org/10.3390/diagnostics10040248