NS1 Recombinant Proteins Are Efficiently Produced in Pichia pastoris and Have Great Potential for Use in Diagnostic Kits for Dengue Virus Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Vectors
2.2. P. pastoris Cloning
2.3. Recombinant NS1-DENV1–4 Protein Production
2.4. NS1-DENV1–4 Purification
2.5. Recombinant Protein Characterization
2.6. Serum Samples
2.7. ELISA
2.8. Statistics
3. Results
3.1. Pichia pastoris NS1-DENV1–4 Cloning
3.2. Expression and Purification of Recombinant Proteins
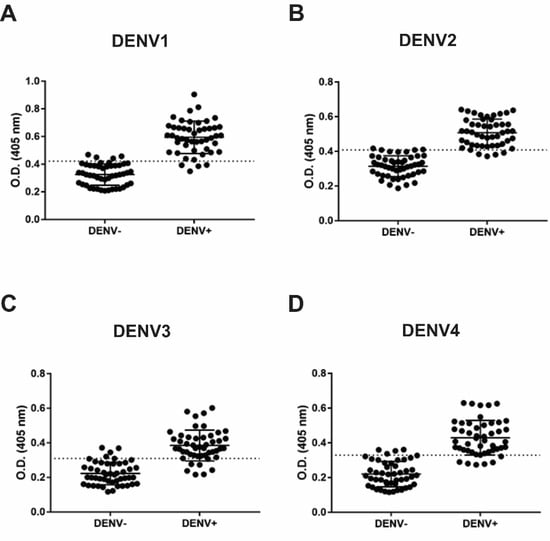
3.3. IgM and IgG Indirect ELISA
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simmons, C.P.; Farrar, J.J.; van Vinh Chau, N.; Wills, B. Dengue. N. Engl. J. Med. 2012, 366, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Mallewa, M. Dengue and other emerging flaviviruses. J. Infect. 2001, 42, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Grard, G.; Moureau, G.; Charrel, R.N.; Holmes, E.C.; Gould, E.A.; de Lamballerie, X. Genomics and evolution of Aedes-borne flaviviruses. J. Gen. Virol. 2010, 91, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Endy, T.P.; Anderson, K.B.; Nisalak, A.; Yoon, I.K.; Green, S.; Rothman, A.L.; Thomas, S.J.; Jarman, R.G.; Libraty, D.H.; Gibbons, R.V. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis. 2011, 5, e975. [Google Scholar] [CrossRef]
- Halstead, S.B. Dengue. Lancet 2007, 370, 1644–1652. [Google Scholar] [CrossRef]
- Caron, M.; Paupy, C.; Grard, G.; Becquart, P.; Mombo, I.; Nso, B.B.; Kassa Kassa, F.; Nkoghe, D.; Leroy, E.M. Recent introduction and rapid dissemination of Chikungunya virus and Dengue virus serotype 2 associated with human and mosquito coinfections in Gabon, central Africa. Clin. Infect. Dis. 2012, 55, e45–e53. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B. Pathogenesis of dengue: Challenges to molecular biology. Science 1988, 239, 476–481. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Guy, B.; Briand, O.; Lang, J.; Saville, M.; Jackson, N. Development of the Sanofi Pasteur tetravalent dengue vaccine: One more step forward. Vaccine 2015, 33, 7100–7111. [Google Scholar] [CrossRef] [Green Version]
- Iacobucci, G. WHO recommends additional tests for Sanofi’s dengue vaccine after safety concerns. BMJ 2018, 361, k1765. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef]
- Brand, C.; Bisaillon, M.; Geiss, B.J. Organization of the Flavivirus RNA replicase complex. Wiley Interdiscip. Rev. RNA 2017, 8, e1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khromykh, A.A.; Sedlak, P.L.; Westaway, E.G. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 2000, 74, 3253–3263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, G.; Randolph, V.B.; Cleaves, G.R.; Ryan, T.E.; Stollar, V. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 1988, 162, 187–196. [Google Scholar] [CrossRef]
- Somnuke, P.; Hauhart, R.E.; Atkinson, J.P.; Diamond, M.S.; Avirutnan, P. N-linked glycosylation of dengue virus NS1 protein modulates secretion, cell-surface expression, hexamer stability, and interactions with human complement. Virology 2011, 413, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Avirutnan, P.; Hauhart, R.E.; Somnuke, P.; Blom, A.M.; Diamond, M.S.; Atkinson, J.P. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 2011, 187, 424–433. [Google Scholar] [CrossRef]
- Young, P.R.; Hilditch, P.A.; Bletchly, C.; Halloran, W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 2000, 38, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Pal, S.; Dauner, A.L.; Mitra, I.; Forshey, B.M.; Garcia, P.; Morrison, A.C.; Halsey, E.S.; Kochel, T.J.; Wu, S.J.L. Evaluation of dengue NS1 antigen rapid tests and ELISA kits using clinical samples. PLoS ONE 2014, 9, e113411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groen, J.; Koraka, P.; Velzing, J.; Copra, C.; Osterhaus, A.D. Evaluation of six immunoassays for detection of dengue virus-specific immunoglobulin M and G antibodies. Clin. Diagn. Lab. Immunol. 2000, 7, 867–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pongsumpun, P.; Garcia Lopez, D.; Favier, C.; Torres, L.; Llosa, J.; Dubois, M.A. Dynamics of dengue epidemics in urban contexts. Trop. Med. Int. Health 2008, 13, 1180–1187. [Google Scholar] [CrossRef]
- Porter, K.R.; Beckett, C.G.; Kosasih, H.; Tan, R.I.; Alisjahbana, B.; Rudiman, P.I.; Widjaja, S.; Listiyaningsih, E.; Ma’roef, C.N.; McArdle, J.L.; et al. Epidemiology of dengue and dengue hemorrhagic fever in a cohort of adults living in Bandung, West Java, Indonesia. Am. J. Trop. Med. Hyg. 2005, 72, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoddard, S.T.; Forshey, B.M.; Morrison, A.C.; Paz-Soldan, V.A.; Vazquez-Prokopec, G.M.; Astete, H.; Reiner, R.C.; Vilcarromero, S.; Elder, J.P.; Halsey, E.S.; et al. House-to-house human movement drives dengue virus transmission. Proc. Natl. Acad. Sci. USA 2013, 110, 994–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. WHO–Dengue Guidelines for Diagnosis, Treatment, Prevention and Control. 2009. Available online: https://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf (accessed on 4 June 2019).
- Pang, J.; Chia, P.Y.; Lye, D.C.; Leo, Y.S. Progress and Challenges towards Point-of-Care Diagnostic Development for Dengue. J. Clin. Microbiol. 2017, 55, 3339–3349. [Google Scholar] [CrossRef]
- Fatima, A.; Wang, J. Review: Progress in the diagnosis of dengue virus infections and importance of point of care test: A review. Pak. J. Pharm. Sci. 2015, 28, 271–280. [Google Scholar]
- Nyan, D.C.; Swinson, K.L. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci. Rep. 2015, 5, 17925. [Google Scholar] [CrossRef] [Green Version]
- Pabbaraju, K.; Wong, S.; Gill, K.; Fonseca, K.; Tipples, G.A.; Tellier, R. Simultaneous detection of Zika, Chikungunya and Dengue viruses by a multiplex real-time RT-PCR assay. J. Clin. Virol. 2016, 83, 66–71. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Gresh, L.; Mohamed-Hadley, A.; Ballesteros, G.; Davila, M.J.; Tellez, Y.; Sahoo, M.K.; Balmaseda, A.; Harris, E.; Pinsky, B.A. Single-Reaction Multiplex Reverse Transcription PCR for Detection of Zika, Chikungunya, and Dengue Viruses. Emerg. Infect. Dis. 2016, 22, 1295–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrar, J.; Focks, D.; Gubler, D.; Barrera, R.; Guzman, M.G.; Simmons, C.; Kalayanarooj, S.; Lum, L.; McCall, P.J.; Lloyd, L.; et al. Towards a global dengue research agenda. Trop. Med. Int. Health 2007, 12, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Gubler, D.J.; Izquierdo, A.; Martinez, E.; Halstead, S.B. Dengue infection. Nat. Rev. Dis. Primers 2016, 2, 16055. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef]
- Nielsen, K.H. Protein expression-yeast. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 536, pp. 133–147. [Google Scholar]
- Schwarzhans, J.-P.; Luttermann, T.; Geier, M.; Kalinowski, J.; Friehs, K. Towards systems metabolic engineering in Pichia pastoris. Biotechnol. Adv. 2017, 35, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Vorauer-Uhl, K.; Lhota, G. Quantification of Recombinant Products in Yeast. In Recombinant Protein Production in Yeast; Humana Press: New York, NY, USA, 2019; pp. 385–428. [Google Scholar]
- Cregg, J.M.; Barringer, K.; Hessler, A.; Madden, K.R. Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 1985, 5, 3376–3385. [Google Scholar] [CrossRef]
- Gasser, B.; Steiger, M.G.; Mattanovich, D. Methanol regulated yeast promoters: Production vehicles and toolbox for synthetic biology. Microb. Cell Factories 2015, 14, 196. [Google Scholar] [CrossRef] [Green Version]
- Puxbaum, V.; Mattanovich, D.; Gasser, B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 2015, 99, 2925–2938. [Google Scholar] [CrossRef]
- Spadiut, O.; Capone, S.; Krainer, F.; Glieder, A.; Herwig, C. Microbials for the production of monoclonal antibodies and antibody fragments. Trends Biotechnol. 2014, 32, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. Fems Microbiol. Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef] [Green Version]
- Weinacker, D.; Rabert, C.; Zepeda, A.B.; Figueroa, C.A.; Pessoa, A.; Farías, J.G. Applications of recombinant Pichia pastoris in the healthcare industry. Braz. J. Microbiol. 2013, 44, 1043–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinhandl, K.; Winkler, M.; Glieder, A.; Camattari, A. Carbon source dependent promoters in yeasts. Microb. Cell Factories 2014, 13, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.L.; Huang, J.H.; Shyu, R.H.; Teng, C.W.; Lin, Y.L.; Kuo, M.D.; Yao, C.W.; Shaio, M.F. High-level expression of recombinant dengue viral NS-1 protein and its potential use as a diagnostic antigen. J. Med Virol. 2001, 65, 553–560. [Google Scholar] [CrossRef]
- Sankar, S.G.; Dhanajeyan, K.J.; Paramasivan, R.; Thenmozhi, V.; Tyagi, B.K.; Vennison, S.J. High-level expression of functionally active Dengue-2 non-structural antigen 1 production in Escherichia coli. Biomed Res. Int. 2013, 2013, 343195. [Google Scholar] [CrossRef] [PubMed]
- Yohan, B.; Wardhani, P.; Trimarsanto, H.; Sasmono, R.T. Production of recombinant dengue non-structural 1 (NS1) proteins from clinical virus isolates. Protein Expr. Purif. 2017, 129, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Landsberg, M.J.; Bletchly, C.; Rothnagel, R.; Waddington, L.; Hankamer, B.; Young, P.R. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J. Gen. Virol. 2012, 93, 771–779. [Google Scholar] [CrossRef]
- Leblois, H.; Young, P.R. Maturation of the dengue-2 virus NS1 protein in insect cells: Effects of downstream NS2A sequences on baculovirus-expressed gene constructs. J. Gen. Virol. 1995, 76, 979–984. [Google Scholar] [CrossRef]
- Allonso, D.; Pereira, I.B.; Alves, A.M.B.; Kurtenbach, E.; Mohana-Borges, R. Expression of soluble, glycosylated and correctly folded dengue virus NS1 protein in Pichia pastoris. Protein Expr. Purif. 2019, 162, 9–17. [Google Scholar] [CrossRef]
- Looke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Kaushik, N.; Rohila, D.; Arora, U.; Raut, R.; Lamminmäki, U.; Khanna, N.; Batra, G. Casamino acids facilitate the secretion of recombinant dengue virus serotype-3 envelope domain III in Pichia pastoris. BMC Biotechnol. 2016, 16, 12. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanaway, J.D.; Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Coffeng, L.E.; Brady, O.J.; Hay, S.I.; Bedi, N.; Bensenor, I.M.; Castañeda-Orjuela, C.A.; et al. The global burden of dengue: An analysis from the Global Burden of Disease Study 2013. Lancet Infect. Dis. 2016, 16, 712–723. [Google Scholar] [CrossRef] [Green Version]
- Peeling, R.W.; Artsob, H.; Pelegrino, J.L.; Buchy, P.; Cardosa, M.J.; Devi, S.; Enria, D.A.; Farrar, J.; Gubler, D.J.; Guzman, M.G.; et al. Evaluation of diagnostic tests: Dengue. Nat. Rev. Microbiol. 2010, 8, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Ooi, E.E. Diagnosis of dengue: An update. Expert Rev. Anti-Infect. Ther. 2012, 10, 895–907. [Google Scholar] [CrossRef]
- Amorim, J.H.; Alves, R.P.; Boscardin, S.B.; Ferreira, L.C. The dengue virus non-structural 1 protein: Risks and benefits. Virus Res. 2014, 181, 53–60. [Google Scholar] [CrossRef]
- Ambrose, J.H.; Sekaran, S.D.; Azizan, A. Dengue Virus NS1 Protein as a Diagnostic Marker: Commercially Available ELISA and Comparison to qRT-PCR and Serological Diagnostic Assays Currently Used by the State of Florida. J. Trop. Med. 2017, 2017, 8072491. [Google Scholar] [CrossRef]
- Libraty, D.H.; Young, P.R.; Pickering, D.; Endy, T.P.; Kalayanarooj, S.; Green, S.; Vaughn, D.W.; Nisalak, A.; Ennis, F.A.; Rothman, A.L. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 2002, 186, 1165–1168. [Google Scholar] [CrossRef]
- Fazlalipour, M.; Keyvani, H.; Monavari, S.H.; Mollaie, H.R. Expression, Purification and Immunogenic Description of a Hepatitis C Virus Recombinant CoreE1E2 Protein Expressed by Yeast Pichia pastoris. Jundishapur J. Microbiol. 2015, 8, e17157. [Google Scholar] [CrossRef] [Green Version]
- Kopera, E.; Dwornyk, A.; Kosson, P.; Florys, K.; Saczynska, V.; Debski, J.; Cecuda-Adamczewska, V.; Szewczyk, B.; Zagórski-Ostoja, W.; Grzelak, K. Expression, purification and characterization of glycosylated influenza H5N1 hemagglutinin produced in Pichia pastoris. Acta Biochim. Pol. 2014, 61, 597–602. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wang, Y. Recent advances in the production of recombinant subunit vaccines in Pichia pastoris. Bioengineered 2016, 7, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Ben Azoun, S.; Belhaj, A.E.; Gongrich, R.; Gasser, B.; Kallel, H. Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris. Microb. Biotechnol. 2016, 9, 355–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Azoun, S.; Ben Zakour, M.; Sghaier, S.; Kallel, H. Expression of rabies virus glycoprotein in the methylotrophic yeast Pichia pastoris. Biotechnol. Appl. Biochem. 2017, 64, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.A.; Paixao, V.F.; Oliveira, M.D.; Honda, E.R.; Oliveira, L.L.; da Silva, C.C.; De Paula, S.O. Dengue-1 envelope protein domain III produced in Pichia pastoris: Potential use for serological diagnosis. Protein Expr. Purif. 2013, 92, 9–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.M.; Jeong, Y.E.; Wang, E.; Lee, Y.J.; Han, M.G.; Park, C.; Lee, W.J.; Choi, W. Cloning and Expression of Recombinant Tick-Borne Encephalitis Virus-like Particles in Pichia pastoris. Osong Public Health Res. Perspect. 2014, 5, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Bawa, Z.; Routledge, S.J.; Jamshad, M.; Clare, M.; Sarkar, D.; Dickerson, I.; Ganzlin, M.; Poyner, D.R.; Bill, R.M. Functional recombinant protein is present in the pre-induction phases of Pichia pastoris cultures when grown in bioreactors, but not shake-flasks. Microb. Cell Factories 2014, 13, 127. [Google Scholar] [CrossRef] [Green Version]
- Rabert, C.; Weinacker, D.; Pessoa, A., Jr.; Farias, J.G. Recombinants proteins for industrial uses: Utilization of Pichia pastoris expression system. Brazilian journal of microbiology. Braz. J. Microbiol. 2013, 44, 351–356. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.C.; Braun-Galleani, S.; Henriquez, M.J.; Bandara, S.; Nesbeth, D. Biotransformation of beta-hydroxypyruvate and glycolaldehyde to l-erythrulose by Pichia pastoris strain GS115 overexpressing native transketolase. Biotechnol. Prog. 2018, 34, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Aw, R.; McKay, P.F.; Shattock, R.J.; Polizzi, K.M. Expressing anti-HIV VRC01 antibody using the murine IgG1 secretion signal in Pichia pastoris. AMB Express 2017, 7, 70. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.C.; Gong, T.; Wang, Q.H.; Liang, X.; Chen, J.J.; Zhu, P. Scaling-up Fermentation of Pichia pastoris to demonstration-scale using new methanol-feeding strategy and increased air pressure instead of pure oxygen supplement. Sci. Rep. 2016, 6, 18439. [Google Scholar] [CrossRef] [Green Version]
- Popov, M.; Li, J.; Reithmeier, R.A. Resolution of glycoproteins by a lectin gel-shift assay. Anal. Biochem. 2000, 279, 90–95. [Google Scholar] [CrossRef]
- Egito, A.S.; Girardet, J.M.; Miclo, L.; Gaillard, J.L. Highly sensitive periodic acid/Schiff detection of bovine milk glycoproteins electrotransferred after nondenaturing electrophoresis, urea electrophoresis, and isoelectric focusing. Le Lait 2001, 81, 775–785. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Rong, Y.; Wang, Y.; Kong, D.; Wang, P.G.; Chen, M.; Kong, Y. Homogeneous production and characterization of recombinant N-GlcNAc-protein in Pichia pastoris. Microb. Cell Factories 2020, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Falgout, B.; Chanock, R.; Lai, C.J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J. Virol. 1989, 63, 1852–1860. [Google Scholar] [CrossRef] [Green Version]
- Falconar, A.; Young, P.; Miles, M.A. Precise location of sequential dengue virus subcomplex and complex B cell epitopes on the nonstructural-1 glycoprotein. Arch. Virol. 1994, 137, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Legge, F.; Lebani, K.; Mahler, S.; Young, P.; Watterson, D.; Treutlein, H.R.; Zeng, J. Computational identification of antibody epitopes on the dengue virus NS1 protein. Molecules 2017, 22, 607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumarasamy, V.; Wahab, A.A.; Chua, S.; Hassan, Z.; Mohamad, M.; Chua, K.B. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J. Virol. Methods 2007, 140, 75–79. [Google Scholar] [CrossRef] [PubMed]






| Abs562 nm | [ ]f (µg/mL) | Yield (mg/L) | |
|---|---|---|---|
| NS1DENV1 | 0.1867 | 1795.0 | 3.590 |
| NS1DENV2 | 0.1790 | 1602.5 | 3.205 |
| NS1DENV3 | 0.1707 | 1395.0 | 2.790 |
| NS1DENV4 | 0.2076 | 2317.5 | 4.635 |
| Standard curve equation: y = 0.0004x + 0.1149; R2 = 0.9963; f: final concentration. | |||
| Anti-IgM | % Sensitivity | % Specificity | p-Value |
|---|---|---|---|
| NS1DENV1 | 91.67 | 91.67 | ≤0.0001 |
| NS1DENV2 | 91.67 | 93.75 | ≤0.0001 |
| NS1DENV3 | 85.42 | 91.67 | ≤0.0001 |
| NS1DENV4 | 87.50 | 91.67 | ≤0.0001 |
| Receiver operating characteristic (ROC) curves were analyzed to estimate the diagnostic sensitivity and specificity. Unpaired t-test for significance. | |||
| Anti-IgG | % Sensitivity | % Specificity | p-Value |
|---|---|---|---|
| NS1DENV1 | 85.42 | 93.75 | ≤0.0001 |
| NS1DENV2 | 87.50 | 91.67 | ≤0.0001 |
| NS1DENV3 | 85.42 | 81.25 | ≤0.0001 |
| NS1DENV4 | 83.33 | 91.67 | ≤0.0001 |
| Receiver operating characteristic (ROC) curves were analyzed to estimate the diagnostic sensitivity and specificity. Unpaired t-test for significance. | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xisto, M.F.; Prates, J.W.O.; Dias, I.M.; Dias, R.S.; da Silva, C.C.; de Paula, S.O. NS1 Recombinant Proteins Are Efficiently Produced in Pichia pastoris and Have Great Potential for Use in Diagnostic Kits for Dengue Virus Infections. Diagnostics 2020, 10, 379. https://doi.org/10.3390/diagnostics10060379
Xisto MF, Prates JWO, Dias IM, Dias RS, da Silva CC, de Paula SO. NS1 Recombinant Proteins Are Efficiently Produced in Pichia pastoris and Have Great Potential for Use in Diagnostic Kits for Dengue Virus Infections. Diagnostics. 2020; 10(6):379. https://doi.org/10.3390/diagnostics10060379
Chicago/Turabian StyleXisto, Mariana Fonseca, John Willians Oliveira Prates, Ingrid Marques Dias, Roberto Sousa Dias, Cynthia Canedo da Silva, and Sérgio Oliveira de Paula. 2020. "NS1 Recombinant Proteins Are Efficiently Produced in Pichia pastoris and Have Great Potential for Use in Diagnostic Kits for Dengue Virus Infections" Diagnostics 10, no. 6: 379. https://doi.org/10.3390/diagnostics10060379
APA StyleXisto, M. F., Prates, J. W. O., Dias, I. M., Dias, R. S., da Silva, C. C., & de Paula, S. O. (2020). NS1 Recombinant Proteins Are Efficiently Produced in Pichia pastoris and Have Great Potential for Use in Diagnostic Kits for Dengue Virus Infections. Diagnostics, 10(6), 379. https://doi.org/10.3390/diagnostics10060379