Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring
Abstract
:1. Introduction
2. Antibiotics
2.1. Classification of Antibiotics
2.1.1. Inhibition of Bacterial Wall Synthesis
2.1.2. Disruption of the Cytoplasmic Membrane
2.1.3. Inhibition of Protein Synthesis
2.1.4. Antibiotics that Act on the Metabolism or Structure of Nucleic Acids
2.1.5. Blocking the Synthesis of Metabolic Factors
2.2. Issue of Bacterial Resistance to Antibiotics
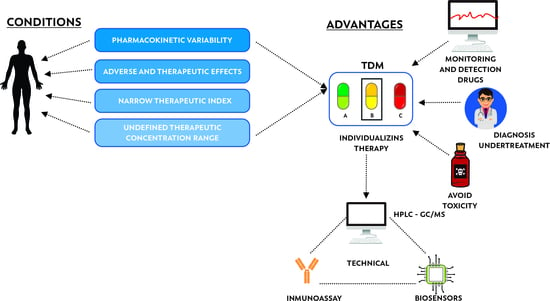
3. Therapeutic Drug Monitoring (TDM)
3.1. Therapeutic Drug Monitoring of Antibiotics as Personalized Medicine
3.2. Antibiotic Quantification Methods
4. Nanobiosensors as Bioanalytic Applications in the Quantification of Antibiotics
4.1. Electrochemical Biosensors
4.2. Optical Biosensors
4.3. Surface-Enhanced Raman Scattering (SERS)
4.4. Piezoelectric (Quartz Crystal Microbalance)
4.5. Nanomechanical Biosensors
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dougherty, T.J.; Pucci, M.J. Antibiotic Discovery and Development; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Chang, H.-H.; Cohen, T.; Grad, Y.H.; Hanage, W.P.; O’Brien, T.F.; Lipsitch, M. Origin and proliferation of multiple-drug resistance in bacterial pathogens. Microbiol. Mol. Biol. Rev. 2015, 79, 101–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, J. The Review on Antimicrobial Resistance Chaired by Jim O’Neill; London; HM Government, Wellcome Trust: London, UK, 2016. [Google Scholar]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Figueras, A. Review of the Evidence to Include TDM in the Essential in Vitro Diagnostics List and Prioritization of Medicines to Be Monitored; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Touw, D.J.; Neef, C.; Thomson, A.H.; Vinks, A.A. Cost-effectiveness of therapeutic drug monitoring: A systematic review. Ther. Drug Monit. 2005, 27, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Reeves, D.; Lovering, A.; Thomson, A. Therapeutic drug monitoring in the past 40 years of the Journal of Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 2016, 71, 3330–3332. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-W. Therapeutic drug monitoring (TDM) of antimicrobial agents. Infect. Chemother. 2008, 40, 133–139. [Google Scholar] [CrossRef]
- Garzon, V.; Pinacho, D.G.; Bustos, R.H.; Garzon, G.; Bustamante, S. Optical Biosensors for Therapeutic Drug Monitoring. Biosensors 2019, 9, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mabilat, C.; Gros, M.F.; Nicolau, D.; Mouton, J.W.; Textoris, J.; Roberts, J.A.; Cotta, M.O.; Van Belkum, A.; Caniaux, I. Diagnostic and medical needs for therapeutic drug monitoring of antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 791–797. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, A. Therapeutic Drug Monitoring: Newer Drugs and Biomarkers; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Horn, D.; Klaas, C.; Fobker, M.; Köck, R.; Lanckohr, C. Therapeutic drug monitoring of antibiotics in critically ill patients. In Handbook of Analytical Separations; Elsevier: Amsterdam, The Netherlands, 2020; Volume 7, pp. 169–183. [Google Scholar]
- Peloquin, C. The role of therapeutic drug monitoring in mycobacterial infections. Tuberc. Nontuberculous Mycobact. Infect. 2017, 5, 119–127. [Google Scholar]
- Meneghello, A.; Tartaggia, S.; Alvau, M.D.; Polo, F.; Toffoli, G. Biosensing technologies for therapeutic drug monitoring. Curr. Med. Chem. 2018, 25, 4354–4377. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.G.; Kim, Y.-G.; Chung, B.G.; Demirci, U.; Khademhosseini, A. Nano/Microfluidics for diagnosis of infectious diseases in developing countries. J. Adv. Drug Deliv. Rev. 2010, 62, 449–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attaallah, R.; Antonacci, A.; Arduini, F.; Amine, A.; Scognamiglio, V. Nanobiosensors for Bioclinical Applications: Pros and Cons. In Green Nanoparticles; Springer: Berlin/Heidelberg, Germany, 2020; pp. 117–149. [Google Scholar]
- Noah, N.M.; Ndangili, P.M. Current Trends of Nanobiosensors for Point-of-Care Diagnostics. J. Anal. Methods Chem. 2019, 2019, 2179718. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, B.D.; Chaubey, A. Biosensors for clinical diagnostics industry. J. Sens. Actuators B Chem. 2003, 91, 117–127. [Google Scholar] [CrossRef]
- Bueno, J. Biosensors in antimicrobial drug discovery: Since biology until screening platforms. J. Microb. Biochem. Technol. 2014, 10. [Google Scholar] [CrossRef] [Green Version]
- D’Orazio, P. Biosensors in clinical chemistry. J. Clin. Chim. Acta 2003, 334, 41–69. [Google Scholar] [CrossRef]
- Korposh, S.; Chianella, I.; Guerreiro, A.; Caygill, S.; Piletsky, S.; James, S.W.; Tatam, R.P. Selective vancomycin detection using optical fibre long period gratings functionalised with molecularly imprinted polymer nanoparticles. Analyst 2014, 139, 2229–2236. [Google Scholar] [CrossRef] [Green Version]
- Carrasco, S.; Benito-Peña, E.; Walt, D.R.; Moreno-Bondi, M.C. Fiber-optic array using molecularly imprinted microspheres for antibiotic analysis. Chem. Sci. 2015, 6, 3139–3147. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wei, Q.; Xu, C.; Li, H.; Wu, D.; Cai, Y.; Mao, K.; Cui, Z.; Du, B. Label-free electrochemical immunosensor for sensitive detection of kanamycin. Sens. Actuators B Chem. 2011, 155, 618–625. [Google Scholar] [CrossRef]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226. [Google Scholar] [CrossRef]
- Calvo, J.; Martínez-Martínez, L. Mecanismos de acción de los antimicrobianos. Enferm. Infecc. Microbiol. Clín. 2009, 27, 44–52. [Google Scholar] [CrossRef]
- Podolsky, S.H. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. 2018, 4, 1–8. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Duin, D.; Paterson, D.L. Multidrug-resistant bacteria in the community: Trends and lessons learned. Infect. Dis. Clin. 2016, 30, 377–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, P.; Paulin, S. The Antibacterial Research and Development Pipeline Needs Urgent Solutions. ACS Infect. Dis. 2020, 6, 1289–1291. [Google Scholar] [CrossRef]
- Genilloud, O. Natural products discovery and potential for new antibiotics. Curr. Opin. Microbiol. 2019, 51, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D. The pharmaceutical industry and the future of drug development. Pharm. Environ. 2015. [Google Scholar] [CrossRef]
- Medina, Á.M. La resistencia a los antibióticos y la falta de interés de la industria farmacéutica. Infectio 2014, 18, 35–36. [Google Scholar] [CrossRef] [Green Version]
- Norrby, S.R.; Nord, C.E.; Finch, R. Lack of development of new antimicrobial drugs: A potential serious threat to public health. Lancet Infect. Dis. 2005, 5, 115–119. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Nielsen, T.B.; Brass, E.P.; Gilbert, D.N.; Bartlett, J.G.; Spellberg, B. Sustainable discovery and development of antibiotics—Is a nonprofit approach the future? N. Engl. J. Med. 2019, 381, 503. [Google Scholar] [CrossRef]
- Hughes, D.; Karlén, A. Discovery and preclinical development of new antibiotics. Upsala J. Med. Sci. 2014, 119, 162–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellberg, B. The future of antibiotics. Crit. Care 2014, 18, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphreys, G.; Fleck, F. United Nations meeting on antimicrobial resistance. Bull. World Health Organ. 2016, 94, 638. [Google Scholar]
- Pacheco, T.; Bustos, R.-H.; González, D.; Garzón, V.; García, J.-C.; Ramírez, D. An approach to measuring colistin plasma levels regarding the treatment of multidrug-resistant bacterial infection. Antibiotics 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacGowan, A.; Macnaughton, E. Antibiotic resistance. Medicine 2017, 45, 622–628. [Google Scholar] [CrossRef]
- Escobar, Q.L. Monitorización terapéutica de fármacos y aspectos prácticos de farmacocinética. Rev. Médica Clínica Las Condes 2016, 27, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2011; pp. 156–167. [Google Scholar]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef]
- Mohr, K.I. History of antibiotics research. In How to Overcome the Antibiotic Crisis; Springer: Berlin/Heidelberg, Germany, 2016; pp. 237–272. [Google Scholar]
- Moore, D. Antibiotic classification and mechanism. J. Retrieved August 2015, 24. [Google Scholar]
- Walsh, C. Antibiotics: Actions, Origins, Resistance; American Society for Microbiology (ASM): Washington, DC, USA, 2003. [Google Scholar]
- Etebu, E.; Arikekpar, I. Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 2016, 4, 90–101. [Google Scholar]
- Pigrau, C. Oxazolidinonas y glucopéptidos. Enfermedades Infecciosas y Microbiologia Clinica 2003, 21, 157–165. [Google Scholar] [CrossRef]
- Garau, M.; Latorre, A.; Alonso-Sanz, M. Fosfomicina: Un antibiótico infravalorado en infecciones urinarias por Escherichia coli. Enfermedades Infecciosas y Microbiologia Clinica 2001, 19, 462–466. [Google Scholar] [CrossRef]
- Johnson, B.A.; Anker, H.; Meleney, F.L. Bacitracin: A new antibiotic produced by a member of the B. subtilis group. Science 1945, 102, 376–377. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Zhu, J.; Zhu, S.; Lu, Y.; Zhang, B.; Lu, K.; Li, J.; Ma, X.; Chen, S. Metabolic engineering of main transcription factors in carbon, nitrogen and phosphorus metabolisms for enhanced production of bacitracin in Bacillus licheniformis. ACS Synth. Biol. 2019, 8, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Sjodt, M.; Brock, K.; Dobihal, G.; Rohs, P.D.; Green, A.G.; Hopf, T.A.; Meeske, A.J.; Srisuknimit, V.; Kahne, D.; Walker, S. Structure and function of the SEDS: bPBP bacterial cell wall synthesis machinery. Found. Crystallogr. 2018, 74, a144. [Google Scholar] [CrossRef]
- Raja, A.; LaBonte, J.; Lebbos, J.; Kirkpatrick, P. Daptomycin. Nat. Rev. Drug Discov. 2003, 2, 943–944. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure--activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef] [Green Version]
- Straus, S.K.; Hancock, R.E.W. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Ward, A.; Campoli-Richards, D.M. Mupirocin. Drugs 1986, 32, 425–444. [Google Scholar] [CrossRef]
- Patel, J.B.; Gorwitz, R.J.; Jernigan, J.A. Mupirocin resistance. Clin. Infect. Dis. 2009, 49, 935–941. [Google Scholar] [CrossRef] [Green Version]
- Ament, P.W.; Jamshed, N.; Horne, J.P. Linezolid: Its Role in the Treatment of Gram-Positive, Drug-Resistance Bacterial Infections. Am. Fam. Physician 2002, 65, 663. [Google Scholar]
- Livermore, D.M. Linezolid in vitro: Mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 2003, 51, ii9–ii16. [Google Scholar] [CrossRef] [PubMed]
- Pualomino, J.; Pachón, J. Aminoglucósidos. Enfermedades Infecciosas y Microbiologia Clinica 2003, 21, 105–115. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drinkovic, D.; Fuller, E.R.; Shore, K.P.; Holland, D.J.; Ellis-Pegler, R. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 2001, 48, 315–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spížek, J.; Řezanka, T. Lincomycin, clindamycin and their applications. Appl. Microbiol. Biotechnol. 2004, 64, 455–464. [Google Scholar] [CrossRef]
- Harms, J.M.; Schlünzen, F.; Fucini, P.; Bartels, H.; Yonath, A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2004, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Minchin, S.D.; Busby, S.J. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012, 66, 125–152. [Google Scholar] [CrossRef] [Green Version]
- Oliphant, C.M.; Green, G.M. Quinolones: A comprehensive review. Am. Fam. Physician 2002, 65, 455–464. [Google Scholar]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef] [Green Version]
- Andriole, V.T. The Quinolones; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Van Bambeke, F.; Michot, J.-M.; Van Eldere, J.; Tulkens, P.M. Quinolones in 2005: An update. Clin. Microbiol. Infect. 2005, 11, 256–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geenes, V.; Chambers, J.; Khurana, R.; Shemer, E.W.; Sia, W.; Mandair, D.; Elias, E.; Marschall, H.-U.; Hague, W.; Williamson, C. Rifampicin in the treatment of severe intrahepatic cholestasis of pregnancy. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 189, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Sveroni, D.; Stefos, A.; Rigopoulou, E.I.; Dalekos, G.N. Rifampicin: Not always an innocent drug. BMJ Case Rep. CP 2018, 11, e227356. [Google Scholar] [CrossRef] [PubMed]
- Dingsdag, S.A.; Hunter, N. Metronidazole: An update on metabolism, structure–cytotoxicity and resistance mechanisms. J. Antimicrob. Chemother. 2017, 73, 265–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, C.W.; Jarrad, A.M.; Cooper, M.A.; Blaskovich, M.A. Nitroimidazoles: Molecular fireworks that combat a broad spectrum of infectious diseases. J. Med. Chem. 2017, 60, 7636–7657. [Google Scholar] [CrossRef] [PubMed]
- Doron, S.I.; Beaulac, K.R.; Dhand, A.; Snydman, D.R. Mechanisms of Resistance in Metronidazole. In Antimicrobial Drug Resistance; Springer: Berlin/Heidelberg, Germany, 2017; pp. 281–287. [Google Scholar]
- Miura, K.; Reckendorf, H. 6 The Nitrofurans. In Progress in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 1967; Volume 5, pp. 320–381. [Google Scholar]
- Grayson, M.L.; Whitby, M. 88 Nitrofurans: Nitrofurazone, Furazolidone and Nitrofurantoin. Use Antibiot. 2010, 1195. [Google Scholar]
- Carta, F.; Scozzafava, A.; Supuran, C.T. Sulfonamides: A patent review (2008–2012). Expert Opin. Ther. Pat. 2012, 22, 747–758. [Google Scholar] [CrossRef]
- Kim, H.U.; Blin, K.; Lee, S.Y.; Weber, T. Recent development of computational resources for new antibiotics discovery. Curr. Opin. Microbiol. 2017, 39, 113–120. [Google Scholar] [CrossRef]
- Juretic, D.; Vukicevic, D.; Ilic, N.; Antcheva, N.; Tossi, A. Computational design of highly selective antimicrobial peptides. J. Chem. Inf. Modeling 2009, 49, 2873–2882. [Google Scholar] [CrossRef]
- Bozhüyük, K.A.; Micklefield, J.; Wilkinson, B. Engineering enzymatic assembly lines to produce new antibiotics. Curr. Opin. Microbiol. 2019, 51, 88–96. [Google Scholar] [CrossRef]
- Migone, T.-S.; Subramanian, G.M.; Zhong, J.; Healey, L.M.; Corey, A.; Devalaraja, M.; Lo, L.; Ullrich, S.; Zimmerman, J.; Chen, A. Raxibacumab for the treatment of inhalational anthrax. N. Engl. J. Med. 2009, 361, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, A.W.; Morrill, A.M. Obiltoxaximab: Adding to the treatment arsenal for Bacillus anthracis infection. Ann. Pharmacother. 2017, 51, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Bezlotoxumab: First global approval. Drugs 2016, 76, 1793–1798. [Google Scholar] [CrossRef] [PubMed]
- Bettiol, E.O.; Harbarth, S.J. Development of new antibiotics: Taking off finally? Swiss Med. Wkly. 2015, 145, w14167. [Google Scholar] [CrossRef]
- Jackson, N.; Czaplewski, L.; Piddock, L.J. Discovery and development of new antibacterial drugs: Learning from experience? J. Antimicrob. Chemother. 2018, 73, 1452–1459. [Google Scholar] [CrossRef] [Green Version]
- Bakkali, M. Could DNA uptake be a side effect of bacterial adhesion and twitching motility? Arch. Microbiol. 2013, 195, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Giedraitienė, A.; Vitkauskienė, A.; Naginienė, R.; Pavilonis, A. Antibiotic resistance mechanisms of clinically important bacteria. Medicina 2011, 47, 19. [Google Scholar] [CrossRef]
- Džidić, S.; Šušković, J.; Kos, B. Antibiotic resistance mechanisms in bacteria: Biochemical and genetic aspects. Food Technol. Biotechnol. 2008, 46, 11–21. [Google Scholar]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Canton, R. Antibiotic resistance genes from the environment: A perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin. Microbiol. Infect. 2009, 15, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.M.; Van Elsas, J.D. 7 Horizontal Gene Transfer. Mod. Soil Microbiol. 2019, 105. [Google Scholar]
- Lerminiaux, N.A.; Cameron, A.D. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- De Oliveira, D.M.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Fleeman, R. Discovering Antibacterial and Anti-Resistance Agents Targeting Multi-Drug Resistant ESKAPE Pathogens. Ph.D. Thesis, University of South Florida, St. Petersburg, FL, USA, 2017. [Google Scholar]
- Singh, S.B.; Young, K.; Silver, L.L. What is an “ideal” antibiotic? Discovery challenges and path forward. Biochem. Pharmacol. 2017, 133, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 2006, 34, S20–S28. [Google Scholar] [CrossRef]
- Kümmerer, K. Drugs in the environment: Emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources—A review. Chemosphere 2001, 45, 957–969. [Google Scholar] [CrossRef]
- Gasilova, N.; Eremin, S. Determination of chloramphenicol in milk by a fluorescence polarization immunoassay. J. Anal. Chem. 2010, 65, 255–259. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Huang, S.-H.; Singco, B.; Huang, H.-Y. Analyses of sulfonamide antibiotics in meat samples by on-line concentration capillary electrochromatography–mass spectrometry. J. Chromatogr. A 2011, 1218, 7640–7647. [Google Scholar] [CrossRef]
- Hamscher, G.; Sczesny, S.; Höper, H.; Nau, H. Determination of persistent tetracycline residues in soil fertilized with liquid manure by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. Anal. Chem. 2002, 74, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Weinberg, H.S.; Meyer, M.T. Trace analysis of trimethoprim and sulfonamide, macrolide, quinolone, and tetracycline antibiotics in chlorinated drinking water using liquid chromatography electrospray tandem mass spectrometry. Anal. Chem. 2007, 79, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Batt, A.L.; Aga, D.S. Simultaneous analysis of multiple classes of antibiotics by ion trap LC/MS/MS for assessing surface water and groundwater contamination. Anal. Chem. 2005, 77, 2940–2947. [Google Scholar] [CrossRef]
- World Health Organization. WHO Global Strategy for Containment of Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE; The University of Chicago Press: Chicago, IL, USA, 2008. [Google Scholar]
- Hamad, B. The antibiotics market. Nat. Rev. Drug Discov. 2010, 9, 675–676. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Deak, D.; Outterson, K.; Powers, J.H.; Kesselheim, A.S. Progress in the fight against multidrug-resistant bacteria? A review of US Food and Drug Administration–approved antibiotics, 2010–2015. Ann. Intern. Med. 2016, 165, 363–372. [Google Scholar] [CrossRef]
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Antibiotic Resistance; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Gabastou, J.-M.; Agudelo, C.I.; Brandileone, M.; Castaneda, E.; Di, J.F. Characterization of invasive isolates of S. pneumoniae, H. influenzae, and N. meningitidis in Latin America and the Caribbean: SIREVA II, 2000–2005. Rev. Panam. Salud Publica PAN Am. J. Public Health 2008, 24, 1–15. [Google Scholar] [CrossRef]
- Younis, W.; AbdelKhalek, A.; S Mayhoub, A.; N Seleem, M. In vitro screening of an FDA-approved library against ESKAPE pathogens. Curr. Pharm. Des. 2017, 23, 2147–2157. [Google Scholar] [CrossRef] [Green Version]
- WHO. High Levels of Antibiotic Resistance Found Worldwide, New Data Shows; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Freire-Moran, L.; Aronsson, B.; Manz, C.; Gyssens, I.C.; So, A.D.; Monnet, D.L.; Cars, O.; Group, E.-E.W. Critical shortage of new antibiotics in development against multidrug-resistant bacteria—Time to react is now. Drug Resist. Updates 2011, 14, 118–124. [Google Scholar] [CrossRef]
- Roberts, J.A.; Norris, R.; Paterson, D.L.; Martin, J.H. Therapeutic drug monitoring of antimicrobials. Br. J. Clin. Pharmacol. 2012, 73, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, I.; Shorten, R.J. Therapeutic drug monitoring of antimicrobials. Ann. Clin. Biochem. 2016, 53, 333–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosso-Fernández, C.; Garnacho-Montero, J.; Antonelli, M.; Dimopoulos, G.; Cisneros, J.M.; Group, M.S. Safety and efficacy of colistin versus meropenem in the empirical treatment of ventilator-associated pneumonia as part of a macro-project funded by the Seventh Framework Program of the European Commission studying off-patent antibiotics: Study protocol for a randomized controlled trial. Trials 2015, 16, 102. [Google Scholar] [PubMed] [Green Version]
- Schmitz, F.-J.; Fluit, A.C.; Gondolf, M.; Beyrau, R.; Lindenlauf, E.; Verhoef, J.; Heinz, H.-P.; Jones, M.E. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 1999, 43, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, L.; Pinilla, G.; Ruiz-Parra, A.I.; Cifuentes, Y.; Gallego, E.A. Determinación del gen aac (6)-aph (2) asociado con resistencia a aminoglucósidos en cepas de Staphylococcus coagulasa negativa en una unidad neonatal en Bogotá. Revista de la Facultad de Medicina 2009, 57, 326–333. [Google Scholar]
- Chow, J.W.; Kak, V.; You, I.; Kao, S.J.; Petrin, J.; Clewell, D.B.; Lerner, S.A.; Miller, G.H.; Shaw, K.J. Aminoglycoside Resistance Genesaph (2″)-Ib and aac (6′)-Im Detected Together in Strains of both Escherichia coli and Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 2691–2694. [Google Scholar] [CrossRef] [Green Version]
- Boehr, D.D.; Farley, A.R.; Wright, G.D.; Cox, J.R. Analysis of the π-π stacking interactions between the aminoglycoside antibiotic kinase APH (3′)-IIIa and its nucleotide ligands. Chem. Biol. 2002, 9, 1209–1217. [Google Scholar] [CrossRef] [Green Version]
- Lambert, T.; Gerbaud, G.; Galimand, M.; Courvalin, P. Characterization of Acinetobacter haemolyticus aac(6′)-Ig gene encoding an aminoglycoside 6′-N-acetyltransferase which modifies amikacin. Antimicrob. Agents Chemother. 1993, 37, 2093–2100. [Google Scholar] [CrossRef] [Green Version]
- Quintiliani, R., Jr.; Evers, S.; Courvalin, P. The van B gene confers various levels of self-transferable resistance to vancomycin in enterococci. J. Infect. Dis. 1993, 167, 1220–1223. [Google Scholar] [CrossRef]
- Evers, S.; Courvalin, P. Regulation of VanB-type vancomycin resistance gene expression by the VanS (B)-VanR (B) two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 1996, 178, 1302–1309. [Google Scholar] [CrossRef] [Green Version]
- Dutka-Malen, S.; Molinas, C.; Arthur, M.; Courvalin, P. Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a D-alanine: D-alanine ligase-related protein necessary for vancomycin resistance. Gene 1992, 112, 53–58. [Google Scholar] [CrossRef]
- Patino, L.A.; Courvalin, P.; Perichon, B. vanE gene cluster of vancomycin-resistant Enterococcus faecalis BM4405. J. Bacteriol. 2002, 184, 6457–6464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perichon, B.; Reynolds, P.; Courvalin, P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 1997, 41, 2016–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depardieu, F.; Bonora, M.G.; Reynolds, P.E.; Courvalin, P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 2003, 50, 931–948. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lin, D.; Yan, G.; Ye, X.; Wu, S.; Guo, Y.; Zhu, D.; Hu, F.; Zhang, Y.; Wang, F. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4643–4647. [Google Scholar] [CrossRef] [Green Version]
- Boyd, D.A.; Willey, B.M.; Fawcett, D.; Gillani, N.; Mulvey, M.R. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel d-Ala-d-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 2008, 52, 2667–2672. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, J.K.; Thomas, L.; Partridge, S.R.; Van Der Reijden, T.; Dijkshoorn, L.; Iredell, J. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 2007, 45, 453–460. [Google Scholar] [CrossRef] [Green Version]
- Khorsi, K.; Messai, Y.; Hamidi, M.; Ammari, H.; Bakour, R. High prevalence of multidrug-resistance in Acinetobacter baumannii and dissemination of carbapenemase-encoding genes blaOXA-23-like, blaOXA-24-like and blaNDM-1 in Algiers hospitals. Asian Pac. J. Trop. Med. 2015, 8, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, S.; Poirel, L.; Papa, A.; Koulourida, V.; Nordmann, P. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob. Agents Chemother. 2009, 53, 4045–4047. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Nordmann, P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 1442–1448. [Google Scholar] [CrossRef] [Green Version]
- Vera-Leiva, A.; Barría-Loaiza, C.; Carrasco-Anabalón, S.; Lima, C.; Aguayo-Reyes, A.; Domínguez, M.; Bello-Toledo, H.; González-Rocha, G. KPC: Klebsiella pneumoniae carbapenemasa, principal carbapenemasa en enterobacterias. Rev. Chil. Infectología 2017, 34, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, A.E.; Stoesser, N.; Wilson, D.J.; Sebra, R.; Kasarskis, A.; Anson, L.W.; Giess, A.; Pankhurst, L.J.; Vaughan, A.; Grim, C.J. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 2016, 60, 3767–3778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, H.; Kim, J.-W.; Kim, J.; Lee, J.H.; Choe, K.W.; Gotoh, N. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2001, 45, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, O.; Juan, C.; Cercenado, E.; Navarro, F.; Bouza, E.; Coll, P.; Pérez, J.; Oliver, A. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 2007, 51, 4329–4335. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Dzink-Fox, J.L.; Chen, M.; Levy, S.B. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: Role ofacrR mutations. Antimicrob. Agents Chemother. 2001, 45, 1515–1521. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Chan, E.; Lam, A.; Cheng, A. Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob. Agents Chemother. 2003, 47, 3567–3573. [Google Scholar] [CrossRef] [Green Version]
- Belland, R.; Morrison, S.; Ison, C.; Huang, W. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 1994, 14, 371–380. [Google Scholar] [CrossRef]
- Arsène, S.; Leclercq, R. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 2007, 51, 3254–3258. [Google Scholar] [CrossRef] [Green Version]
- Strahilevitz, J.; Engelstein, D.; Adler, A.; Temper, V.; Moses, A.E.; Block, C.; Robicsek, A. Changes in qnr prevalence and fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae and Enterobacter spp. collected from 1990 to 2005. Antimicrob. Agents Chemother. 2007, 51, 3001–3003. [Google Scholar] [CrossRef] [Green Version]
- Morales, G.; Picazo, J.J.; Baos, E.; Candel, F.J.; Arribi, A.; Peláez, B.; Andrade, R.; De la Torre, M.-Á.; Fereres, J.; Sánchez-García, M. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 2010, 50, 821–825. [Google Scholar] [CrossRef] [Green Version]
- Marshall, S.; Donskey, C.; Hutton-Thomas, R.; Salata, R.; Rice, L. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellido, J.L.M. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev. Esp. Quimioter. 2017, 30, 391–396. [Google Scholar]
- Rincón, S.; Panesso, D.; Díaz, L.; Carvajal, L.P.; Reyes, J.; Munita, J.M.; Arias, C.A. Resistencia a antibióticos de última línea en cocos Gram positivos: La era posterior a la vancomicina. Biomedica Revista del Instituto Nacional de Salud 2014, 34, 191. [Google Scholar]
- Nummila, K.; Kilpeläinen, I.; Zähringer, U.; Vaara, M.; Helander, I.M. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coii are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 1995, 16, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-S.; Liu, M.-C.; Teng, L.-J.; Wang, W.-B.; Hsueh, P.-R.; Liaw, S.-J. Proteus mirabilis pmrI, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation, and urothelial cell invasion. Antimicrob. Agents Chemother. 2010, 54, 1564–1571. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.O.; Dia, N.M.; Gautret, P.; Benkouiten, S.; Belhouchat, K.; Drali, T.; Parola, P.; Brouqui, P.; Memish, Z.; Raoult, D. Acquisition of extended-spectrum cephalosporin-and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int. J. Antimicrob. Agents 2015, 45, 600–604. [Google Scholar] [CrossRef]
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.-V.; Yilmaz, M.; Nordmann, P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766. [Google Scholar] [CrossRef] [Green Version]
- Jaidane, N.; Bonnin, R.A.; Mansour, W.; Girlich, D.; Creton, E.; Cotellon, G.; Chaouch, C.; Boujaafar, N.; Bouallegue, O.; Naas, T. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob. Agents Chemother. 2018, 62, e01601–e01617. [Google Scholar] [CrossRef] [Green Version]
- Diao, L.; Meibohm, B. Pharmacokinetics and pharmacokinetic–pharmacodynamic correlations of therapeutic peptides. Clin. Pharmacokinet. 2013, 52, 855–868. [Google Scholar] [CrossRef]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. J. Infect. Drug Resist. 2019, 12, 965. [Google Scholar] [CrossRef] [Green Version]
- Markou, N.; Fousteri, M.; Markantonis, S.L.; Zidianakis, B.; Hroni, D.; Boutzouka, E.; Baltopoulos, G. Colistin pharmacokinetics in intensive care unit patients on continuous venovenous haemodiafiltration: An observational study. J. Antimicrob. Chemother. 2012, 67, 2459–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghiculescu, R. Therapeutic drug monitoring: Which drugs, why, when and how to do it. Aust. Prescr. 2008, 31, 42–44. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-S.; Lee, M.-H. Overview of therapeutic drug monitoring. Korean J. Intern. Med. 2009, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Biran, I.; Rissin, D.M.; Ron, E.Z.; Walt, D.R. Optical imaging fiber-based live bacterial cell array biosensor. Anal. Biochem. 2003, 315, 106–113. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; McCarthy, J. Personalized medicine: Revolutionizing drug discovery and patient care. Trends Biotechnol. 2001, 19, 491–496. [Google Scholar] [CrossRef]
- Jain, K.K.; Jain, K. Textbook of Personalized Medicine; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinks, A.A.; Derendorf, H.; Mouton, J.W. Fundamentals of Antimicrobial Pharmacokinetics and Pharmacodynamics; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Dafale, N.A.; Semwal, U.P.; Rajput, R.K.; Singh, G. Selection of appropriate analytical tools to determine the potency and bioactivity of antibiotics and antibiotic resistance. J. Pharm. Anal. 2016, 6, 207–213. [Google Scholar] [CrossRef] [Green Version]
- McKeating, K.S.; Aubé, A.; Masson, J.-F. Biosensors and nanobiosensors for therapeutic drug and response monitoring. Analyst 2016, 141, 429–449. [Google Scholar] [CrossRef]
- Hodiamont, C.J.; Janssen, J.M.; De Jong, M.D.; Mathôt, R.A.; Juffermans, N.P.; Van Hest, R.M. Therapeutic drug monitoring of gentamicin peak concentrations in critically ill patients. Ther. Drug Monit. 2017, 39, 522–530. [Google Scholar] [CrossRef]
- Bamgbola, O. Review of vancomycin-induced renal toxicity: An update. Ther. Adv. Endocrinol. Metab. 2016, 7, 136–147. [Google Scholar] [CrossRef]
- Zasowski, E.J.; Murray, K.P.; Trinh, T.D.; Finch, N.A.; Pogue, J.M.; Mynatt, R.P.; Rybak, M.J. Identification of vancomycin exposure-toxicity thresholds in hospitalized patients receiving intravenous vancomycin. Antimicrob. Agents Chemother. 2018, 62, e01684-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luque, Y.; Louis, K.; Jouanneau, C.; Placier, S.; Esteve, E.; Bazin, D.; Rondeau, E.; Letavernier, E.; Wolfromm, A.; Gosset, C. Vancomycin-associated cast nephropathy. J. Am. Soc. Nephrol. 2017, 28, 1723–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.K.; Mulieri, K.M.; Ishmael, F.T. Characterization of vancomycin reactions and linezolid utilization in the pediatric population. J. Allergy Clin. Immunol. Pract. 2017, 5, 750–756. [Google Scholar] [CrossRef]
- Pea, F.; Brollo, L.; Viale, P.; Pavan, F.; Furlanut, M. Teicoplanin therapeutic drug monitoring in critically ill patients: A retrospective study emphasizing the importance of a loading dose. J. Antimicrob. Chemother. 2003, 51, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, N.; Hu, S.; Xie, J.; Lei, J.e.; Wang, Y.; Zheng, X.; Xing, J.; Dong, Y. Factors on trough teicoplanin levels, associations between levels, efficacy and safety in patients with gram-positive infections. Int. J. Clin. Pharmacol. Ther. 2015, 53, 356–362. [Google Scholar] [CrossRef]
- Grensemann, J.; Busse, D.; König, C.; Roedl, K.; Jäger, W.; Jarczak, D.; Iwersen-Bergmann, S.; Manthey, C.; Kluge, S.; Kloft, C. Acute-on-chronic liver failure alters meropenem pharmacokinetics in critically ill patients with continuous hemodialysis: An observational study. Ann. Intensive Care 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Ehmann, L.; Zoller, M.; Minichmayr, I.K.; Scharf, C.; Maier, B.; Schmitt, M.V.; Hartung, N.; Huisinga, W.; Vogeser, M.; Frey, L. Role of renal function in risk assessment of target non-attainment after standard dosing of meropenem in critically ill patients: A prospective observational study. Crit. Care 2017, 21, 263. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.S.; Machado, A.S.; Mendes, E.T.; Chaves, L.; Neto, L.V.P.; Da Silva Jr, C.V.; Santos, S.R.C.J.; Sanches, C.; Macedo, E.; Levin, A.S. Pharmacokinetic and Pharmacodynamic Characteristics of Vancomycin and Meropenem in Critically Ill Patients Receiving Sustained Low-Efficiency Dialysis. Clin. Ther. 2020, 42, 625–633. [Google Scholar] [CrossRef]
- Rapp, M.; Urien, S.; Foissac, F.; Béranger, A.; Bouazza, N.; Benaboud, S.; Bille, E.; Zheng, Y.; Gana, I.; Moulin, F. Population pharmacokinetics of meropenem in critically ill children with different renal functions. Eur. J. Clin. Pharmacol. 2020, 76, 61–71. [Google Scholar] [CrossRef]
- Vardakas, K.Z.; Voulgaris, G.L.; Maliaros, A.; Samonis, G.; Falagas, M.E. Prolonged versus short-term intravenous infusion of antipseudomonal β-lactams for patients with sepsis: A systematic review and meta-analysis of randomised trials. Lancet Infect. Dis. 2018, 18, 108–120. [Google Scholar] [CrossRef]
- Lipsky, B.A.; Baker, C.A. Fluoroquinolone toxicity profiles: A review focusing on newer agents. Clin. Infect. Dis. 1999, 28, 352–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barlow, J.L.; Mitchon, G.J. Fluoroquinolone Toxicity Mimicking Septic Shock in an Elderly Male. In B56. Critical Care Case Reports: ICU Toxicology; American Thoracic Society: New York, NY, USA, 2017; p. A3782. [Google Scholar]
- Murtaza, G.; Boonpheng, B. Fluoroquinolone-Associated Muscle Tear and Hematoma: A Case Report. Am. J. Ther. 2018, 25, e386–e388. [Google Scholar] [CrossRef] [PubMed]
- Patterson, D.R. Quinolone toxicity: Methods of assessment. Am. J. Med. 1991, 91, S35–S37. [Google Scholar] [CrossRef]
- Pea, F.; Furlanut, M.; Cojutti, P.; Cristini, F.; Zamparini, E.; Franceschi, L.; Viale, P. Therapeutic drug monitoring of linezolid: A retrospective monocentric analysis. Antimicrob. Agents Chemother. 2010, 54, 4605–4610. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, Y.; Yukawa, E.; Hiraki, Y.; Matsumoto, K.; Mizoguchi, A.; Morita, K.; Kamimura, H.; Karube, Y.; To, H. Population pharmacokinetic analysis of linezolid in low body weight patients with renal dysfunction. J. Clin. Pharmacol. 2013, 53, 967–973. [Google Scholar] [CrossRef]
- Nukui, Y.; Hatakeyama, S.; Okamoto, K.; Yamamoto, T.; Hisaka, A.; Suzuki, H.; Yata, N.; Yotsuyanagi, H.; Moriya, K. High plasma linezolid concentration and impaired renal function affect development of linezolid-induced thrombocytopenia. J. Antimicrob. Chemother. 2013, 68, 2128–2133. [Google Scholar] [CrossRef] [Green Version]
- Tobias, P.E.; Varughese, C.A.; Hanson, A.P.; Gurnani, P.K. A case of linezolid induced toxicity. J. Pharm. Pract. 2020, 33, 222–225. [Google Scholar] [CrossRef]
- Garrabou, G.; Soriano, A.; Pinos, T.; Casanova-Molla, J.; Pacheu-Grau, D.; Moren, C.; Garcia-Arumi, E.; Morales, M.; Ruiz-Pesini, E.; Catalan-Garcia, M. Mitochondrial toxicity of linezolid in blood cells and skin nerve fibers: Influence of mitochondrial genetics. Antimicrob. Agents Chemother. 2017. [Google Scholar] [CrossRef] [Green Version]
- Ellis, M.K.; Szabo, A.; Joshi, K. Baseline Characteristics and Demographics of Patients Receiving Daptomycin and Incidence of Toxicity Associated with Daptomycin Use. In A44. Drug Induced and Rare Lung Disease; American Thoracic Society: New York, NY, USA, 2016; p. A1591. [Google Scholar]
- Kido, K.; Oyen, A.A.; Beckmann, M.A.; Brouse, S.D. Musculoskeletal toxicities in patients receiving concomitant statin and daptomycin therapy. Am. J. Health-Syst. Pharm. 2019, 76, 206–210. [Google Scholar] [CrossRef]
- Wasko, J.A.; Dietrich, E.; Davis, K. Risk of Daptomycin-associated Myopathy with Concomitant Statin Use. Clin. Infect. Dis. 2018, 69, 558–559. [Google Scholar] [CrossRef]
- Gao, X.; Bachan, M.; Khan, Z.; Siegel, R. Daptomycin Induced Rhabdomyolysis: A Rare Complication in Critical Care Patients. In D46. Critical Care Case Reports: Toxicology and Poisonings; American Thoracic Society: New York, NY, USA, 2018; p. A6918. [Google Scholar]
- Janda, A.; Jogendra, M.R. A case report and literature review of daptomycin-induced liver injury. IDCases 2018, 14, e00452. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Arslan, A.; Atiq, M.U.; Chan, V.; Patel, R.K. Unexpected Outcome of Daptomycin-Induced Eosinophilic Pneumonia: Rarity within a Rarity. Cureus 2019, 11, e6271. [Google Scholar] [CrossRef] [PubMed]
- Spapen, H.; Jacobs, R.; Van Gorp, V.; Troubleyn, J.; Honoré, P.M. Renal and neurological side effects of colistin in critically ill patients. Ann. Intensive Care 2011, 1, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nation, R.L.; Garonzik, S.M.; Li, J.; Thamlikitkul, V.; Giamarellos-Bourboulis, E.J.; Paterson, D.L.; Turnidge, J.D.; Forrest, A.; Silveira, F. Updated US and European dose recommendations for intravenous colistin: How do they perform? Clin. Infect. Dis. 2016, 62, 552–558. [Google Scholar] [CrossRef] [Green Version]
- Danhof, M.; Breimer, D. Therapeutic drug monitoring in saliva. Clin. Pharmacokinet. 1978, 3, 39–57. [Google Scholar] [CrossRef]
- Pincus, M.R.; Bluth, M.H.; Abraham, N.Z., Jr. Toxicology and therapeutic drug monitoring. In Henry’s Clinical Diagnosis and Management by Laboratory Methods E-Book; Elsevier: Amsterdam, The Netherlands, 2017; p. 324. [Google Scholar]
- Castro-Orozco, R.; Barreto-Maya, A.C.; Guzmán-Álvarez, H.; Ortega-Quiroz, R.J.; Benítez-Peña, L. Antimicrobial resistance pattern for gram-negative uropathogens isolated from hospitalised patients and outpatients in Cartagena, 2005–2008. Rev. Salud Publica 2010, 12, 1010–1019. [Google Scholar] [CrossRef] [Green Version]
- García Domínguez, M.E. Monitorización Terapéutica de Fármacos. Una Visión General; Universidad de Sevilla: Sevilla, Spain, 2018. [Google Scholar]
- Randjelovic, P.; Veljkovic, S.; Stojiljkovic, N.; Sokolovic, D.; Ilic, I. Gentamicin nephrotoxicity in animals: Current knowledge and future perspectives. EXCLI J. 2017, 16, 388. [Google Scholar]
- De Gatta, M.F.; Mendez, M.; Romano, S.; Calvo, M.; Dominguez-Gil, A.; Lanao, J. Pharmacokinetics of amikacin in intensive care unit patients. J. Clin. Pharm. Ther. 1996, 21, 417–421. [Google Scholar] [CrossRef]
- Zaheer, Z.; Chiragh, S. Evaluation of Safety of Tobramycin. Biomedica 2017, 22, 110–116. [Google Scholar]
- Domenech, A. Estudio Experimental de la Eficacia de los Glucopéptidos en Monoterapia o Con B-Lactámicos en la Infección por Staphylococcus aureus con Sensibilidad Disminuida a Glucopéptidos; Universidad de Barcelona: Barcelona, Spain, 2006. [Google Scholar]
- Lestner, J.M.; Hill, L.F.; Heath, P.T.; Sharland, M. Vancomycin toxicity in neonates: A review of the evidence. Curr. Opin. Infect. Dis. 2016, 29, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Boyd, A.; Brasa, C.C. Teicoplanin therapeutic drug monitoring (TDM)–excessive or essential? Access Microbiol. 2020, 2, 213. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kang, C.-I.; Huh, K.; Cho, S.Y.; Chung, D.R.; Lee, S.-Y.; Kim, Y.-J.; Peck, K.R. Evaluating the optimal dose of teicoplanin with therapeutic drug monitoring: Not too high for adverse event, not too low for treatment efficacy. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Luque, S.; Grau, S.; Berenguer, N.; Horcajada, J.P.; Sorlí, L.; Montero, M.M.; Salas, E. Luces y sombras en el uso de colistina: Falta mucho por conocer. Enferm. Infecc. Y Microbiol. Clínica 2011, 29, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Barcelona, L.; Marin, M.; Stamboulian, D. Betalactámicos con inhibidores de betalactamasas: Amoxicilina-sulbactam. Medicina 2008, 68, 65–74. [Google Scholar] [PubMed]
- Arango, C.; Villamarin, N.; Gallardo, L.M.; De Alviz, A.L.; De Ramos, M.E.; De Mejía, L.A. Tres generaciones de cefalosporinas. Estructura, farmacología y actividad antimicrobiana. Biomédica 1985, 5, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Andraca Perera, J.R.; Rodríguez Gil, E.; Fundora Santana, A. Cefalosporinas. Rev. Cuba. Farm. 2001, 35, 219–222. [Google Scholar]
- Rivas, K.; Rivas, M.; Dávila, E.; Rodríguez, M. Cefalosporinas: De la primera a la cuarta generación. Rev. Fac. Med. 2002, 25, 142–153. [Google Scholar]
- González-Ruiz, A.; de León, S.P.; Ruiz-Palacios, G. Cefalosporinas de tercera generación: Las dos caras de la moneda. Salud Pública México 2014, 27, 479–484. [Google Scholar]
- Martínez, M.J.F.; García, M.I.G.; Sánchez, E.G.; Sánchez, J.E.G. Los carbapenems disponibles: Propiedades y diferencias. Enfermedades Infecciosas y Microbiología Clínica 2010, 28, 53–64. [Google Scholar]
- Kumagai, T.; Tamai, S.; Abe, T.; Hikda, M. Current status of oral carbapenem development. Curr. Med. Chem. Anti-Infect. Agents 2002, 1, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Alos, J.-I. Quinolonas. Enferm. Infecc. Microbiol. Clin. 2009, 27, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, A.M.; Jones, A.M. The quinolones: Decades of development and use. J. Antimicrob. Chemother. 2003, 51, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.I.; MacGowan, A.P. Development of the quinolones. J. Antimicrob. Chemother. 2003, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Casparian, J.M.; Luchi, M.; Moffat, R.E.; Hinthorn, D. Quinolones and tendon ruptures. South. Med. J. 2000, 93, 488–491. [Google Scholar] [CrossRef]
- Stahlmann, R.; Lode, H. Toxicity of quinolones. Drugs 1999, 58, 37–42. [Google Scholar] [CrossRef]
- Tomé, A.M.; Filipe, A. Quinolones. Drug Saf. 2011, 34, 465–488. [Google Scholar] [CrossRef]
- MacGowan, A.P. Pharmacokinetic and pharmacodynamic profile of linezolid in healthy volunteers and patients with Gram-positive infections. J. Antimicrob. Chemother. 2003, 51, ii17–ii25. [Google Scholar] [CrossRef]
- Moraza, L.; Leache, L.; Aquerreta, I.; Ortega, A. Toxicidad hematológica inducida por linezolid. Farm. Hosp. 2015, 39, 320–332. [Google Scholar]
- Abou Hassan, O.K.; Karnib, M.; El-Khoury, R.; Nemer, G.; Ahdab-Barmada, M.; BouKhalil, P. Linezolid Toxicity and Mitochondrial Susceptibility: A Novel Neurological Complication in a Lebanese Patient. Front. Pharmacol. 2016, 7, 325. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.C.; Vaduganathan, M.; Phillips, K.M.; O’Donnell, W.J. A triad of linezolid toxicity: Hypoglycemia, lactic acidosis, and acute pancreatitis. In Baylor University Medical Center Proceedings; Taylor & Francis: Abingdon, UK, 2015; pp. 466–468. [Google Scholar]
- Araos, R.; García, P.; Chanqueo, L.; Labarca, J. Daptomicina: Características farmacológicas y aporte en el tratamiento de infecciones por cocáceas gram positivas. Rev. Chil. Infectología 2012, 29, 127–131. [Google Scholar] [CrossRef] [Green Version]
- McWhinney, B.C.; Wallis, S.C.; Hillister, T.; Roberts, J.A.; Lipman, J.; Ungerer, J.P. Analysis of 12 beta-lactam antibiotics in human plasma by HPLC with ultraviolet detection. J. Chromatogr. B 2010, 878, 2039–2043. [Google Scholar] [CrossRef] [PubMed]
- Schlüsener, M.P.; Bester, K.; Spiteller, M. Determination of antibiotics such as macrolides, ionophores and tiamulin in liquid manure by HPLC–MS/MS. Anal. Bioanal. Chem. 2003, 375, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Farin, D.; Piva, G.A.; Gozlan, I.; Kitzes-Cohen, R. A modified HPLC method for the determination of vancomycin in plasma and tissues and comparison to FPIA (TDX). J. Pharm. Biomed. Anal. 1998, 18, 367–372. [Google Scholar] [CrossRef]
- Tobin, C.; Darville, J.; Lovering, A.; Macgowan, A. An HPLC assay for daptomycin in serum. J. Antimicrob. Chemother. 2008, 62, 1462–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morovján, G.; Csokan, P.; Nemeth-Konda, L. HPLC determination of colistin and aminoglycoside antibiotics in feeds by post-column derivatization and fluorescence detection. Chromatographia 1998, 48, 32–36. [Google Scholar] [CrossRef]
- Mendez, A.S.; Steppe, M.; Schapoval, E.E. Validation of HPLC and UV spectrophotometric methods for the determination of meropenem in pharmaceutical dosage form. J. Pharm. Biomed. Anal. 2003, 33, 947–954. [Google Scholar] [CrossRef]
- Herregodts, J.; Van Vooren, S.; Deschuyteneer, E.; Dhaese, S.; Stove, V.; Verstraete, A.; De Waele, J. Measuring antibiotics in exhaled air in critically ill, non-ventilated patients: A feasibility and proof of concept study. J. Crit. Care 2019, 51, 46–50. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [Green Version]
- Odekerken, J.C.; Logister, D.M.; Assabre, L.; Arts, J.J.; Walenkamp, G.H.; Welting, T.J. ELISA-based detection of gentamicin and vancomycin in protein-containing samples. SpringerPlus 2015, 4, 614. [Google Scholar] [CrossRef] [Green Version]
- Shanin, I.; Shaimardanov, A.; Thai, N.T.D.; Eremin, S. Determination of fluoroquinolone antibiotic levofloxacin in urine by fluorescence polarization immunoassay. J. Anal. Chem. 2015, 70, 712–717. [Google Scholar] [CrossRef]
- Kitagawa, T.; Ohtani, W.; Maeno, Y.; Fujiwara, K.; Kimura, Y. Sensitive Enzyme Immunoassay of Colistin and Its Application to Detect Residual Colistin in Rainbow Itout Tissue. J. Assoc. Off. Anal. Chem. 1985, 68, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Hammer, P. ELISA zum nachweis von colistin aus rohmilch. Arb. Des. Arb. Lebensm. 1998, 39, 411–414. [Google Scholar]
- Dijkstra, J.; Voerman, A.; Greijdanus, B.; Touw, D.; Alffenaar, J. Immunoassay analysis of kanamycin in serum using the tobramycin kit. Antimicrob. Agents Chemother. 2016, 60, 4646–4651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, D.; Silva, C.; Andresen, M.; Soto, N.; Wong, K.-Y.; Andresen, M. Monitorización terapéutica de antibióticos: Nuevas metodologías: Biosensores. Rev. Médica Chile 2015, 143, 1050–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouws, J.; Van Egmond, H.; Loeffen, G.; Schouten, J.; Keukens, H.; Smulders, I.; Stegeman, H. Suitability of the Charm HVS and a microbiological multiplate system for detection of residues in raw milk at EU maximum residue levels. Vet. Q. 1999, 21, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Van Eenennaam, A.; Cullor, J.S.; Perani, L.; Gardner, I.; Smith, W.; Dellinger, J.; Guterbock, W.; Jensen, L. Evaluation of milk antibiotic residue screening tests in cattle with naturally occurring clinical mastitis. J. Dairy Sci. 1993, 76, 3041–3053. [Google Scholar] [CrossRef]
- Charm, S.E.; Chi, R. Microbial Receptor Assay for Rapi d Detection and Identification of Seven Families of Antimicrobial Drugs in Milk: Collaborative Study. J. Assoc. Off. Anal. Chem. 1988, 71, 304–316. [Google Scholar] [CrossRef]
- Mascini, M.; Tombelli, S. Biosensors for biomarkers in medical diagnostics. Biomarkers 2008, 13, 637–657. [Google Scholar] [CrossRef]
- Losoya-Leal, A.; Estevez, M.-C.; Martínez-Chapa, S.O.; Lechuga, L.M. Design of a surface plasmon resonance immunoassay for therapeutic drug monitoring of amikacin. Talanta 2015, 141, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Jianrong, C.; Yuqing, M.; Nongyue, H.; Xiaohua, W.; Sijiao, L. Nanotechnology and biosensors. Biotechnol. Adv. 2004, 22, 505–518. [Google Scholar] [CrossRef]
- Neumann, T.; Junker, H.; Schmidt, K.; Sekul, R. SPR-based fragment screening: Advantages and applications. Curr. Top. Med. Chem. 2007, 7, 1630–1642. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, L.; Li, F. Mechanistic challenges and advantages of biosensor miniaturization into the nanoscale. ACS Sens. 2017, 2, 458–467. [Google Scholar] [CrossRef]
- Turner, A.; Karube, I.; Wilson, G.S. Biosensors: Fundamentals and Applications; Oxford University Press: Oxford, UK, 1987. [Google Scholar]
- Cass, A.E.; Cooper, J.M. Biosensors: A Practical Approach; IRL Press: Oxford, UK, 1990. [Google Scholar]
- Borisov, S.M.; Wolfbeis, O.S. Optical biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef] [PubMed]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumayor, V.G.; Iglesias, E.G.; Galán, O.R.; Cabezas, L.G. Aplicaciones de Biosensores en la Industria Agroalimentaria; Informes de Vigilancia Tecnológica: Madrid, Spain, 2005. [Google Scholar]
- Bustos Cruz, R.H.; Sanchez, M.M.; Dominguez-Sanchez, M.A.; Barreto, G.E.; Lancheros, D.; Reynolds, J. Nanotechnology in Neurosciences: An Approach. Curr. Pharm. Des. 2017, 23, 4154–4169. [Google Scholar] [CrossRef] [PubMed]
- Vo-Dinh, T.; Cullum, B. Biosensors and biochips: Advances in biological and medical diagnostics. Fresenius’ J. Anal. Chem. 2000, 366, 540–551. [Google Scholar] [CrossRef]
- Reder-Christ, K.; Bendas, G. Biosensor applications in the field of antibiotic research—A review of recent developments. Sensors 2011, 11, 9450–9466. [Google Scholar] [CrossRef] [Green Version]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wright, G.; Yang, Y. Materials and techniques for electrochemical biosensor design and construction. Biosens. Bioelectron. 2000, 15, 273–282. [Google Scholar] [CrossRef]
- Trivedi, U.; Lakshminarayana, D.; Kothari, I.; Patel, N.; Kapse, H.; Makhija, K.; Patel, P.; Panchal, C. Potentiometric biosensor for urea determination in milk. Sens. Actuators B Chem. 2009, 140, 260–266. [Google Scholar] [CrossRef]
- Gomes, S.; Nogueira, J.; Rebelo, M. An amperometric biosensor for polyphenolic compounds in red wine. Biosens. Bioelectron. 2004, 20, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, J.V.; Ahmed, A.; Griffiths, H.H.; Pollock, N.M.; Hooper, N.M.; Millner, P.A. A label-free electrical impedimetric biosensor for the specific detection of Alzheimer’s amyloid-beta oligomers. Biosens. Bioelectron. 2014, 56, 83–90. [Google Scholar] [CrossRef]
- Rowe, A.A.; Miller, E.A.; Plaxco, K.W. Reagentless measurement of aminoglycoside antibiotics in blood serum via an electrochemical, ribonucleic acid aptamer-based biosensor. Anal. Chem. 2010, 82, 7090–7095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, L.M.; Callera, W.F.; Sotomayor, M.D.; Bueno, P.R. Penicillinase-based amperometric biosensor for penicillin G. Electrochem. Commun. 2014, 38, 131–133. [Google Scholar] [CrossRef]
- Blanchaert, B.; Jorge, E.P.; Jankovics, P.; Adams, E.; Van Schepdael, A. Assay of kanamycin A by HPLC with direct UV detection. Chromatographia 2013, 76, 1505–1512. [Google Scholar] [CrossRef]
- Xu, J.-Z.; Zhu, J.-J.; Wang, H.; Chen, H.-Y. Nano-sized copper oxide modified carbon paste electrodes as an amperometric sensor for amikacin. Anal. Lett. 2003, 36, 2723–2733. [Google Scholar] [CrossRef]
- Baietto, L.; D’Avolio, A.; De Rosa, F.G.; Garazzino, S.; Michelazzo, M.; Ventimiglia, G.; Siccardi, M.; Simiele, M.; Sciandra, M.; Di Perri, G. Development and validation of a simultaneous extraction procedure for HPLC-MS quantification of daptomycin, amikacin, gentamicin, and rifampicin in human plasma. Anal. Bioanal. Chem. 2010, 396, 791–798. [Google Scholar] [CrossRef]
- Pinacho, D.G.; Sánchez-Baeza, F.; Pividori, M.-I.; Marco, M.-P. Electrochemical detection of fluoroquinolone antibiotics in milk using a magneto immunosensor. Sensors 2014, 14, 15965–15980. [Google Scholar] [CrossRef] [Green Version]
- González-Fernández, E.; De-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Impedimetric aptasensor for tobramycin detection in human serum. Biosens. Bioelectron. 2011, 26, 2354–2360. [Google Scholar] [CrossRef]
- Shou, D.; Dong, Y.; Shen, L.; Wu, R.; Zhang, Y.; Zhang, C.; Zhu, Y. Rapid quantification of tobramycin and vancomycin by UPLC–TQD and application to osteomyelitis patient samples. J. Chromatogr. Sci. 2014, 52, 501–507. [Google Scholar] [CrossRef] [Green Version]
- Ionescu, R.E.; Jaffrezic-Renault, N.; Bouffier, L.; Gondran, C.; Cosnier, S.; Pinacho, D.G.; Marco, M.-P.; Sánchez-Baeza, F.J.; Healy, T.; Martelet, C. Impedimetric immunosensor for the specific label free detection of ciprofloxacin antibiotic. Biosens. Bioelectron. 2007, 23, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Krol, G.; Beck, G.; Benham, T. HPLC analysis of ciprofloxacin and ciprofloxacin metabolites in body fluids. J. Pharm. Biomed. Anal. 1995, 14, 181–190. [Google Scholar] [CrossRef]
- Paniel, N.; Istamboulié, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T.J.T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, W.; Pei, M.; Ding, F. GR–Fe 3 O 4 NPs and PEDOT–AuNPs composite based electrochemical aptasensor for the sensitive detection of penicillin. Anal. Methods 2016, 8, 4391–4397. [Google Scholar] [CrossRef]
- Jahanbani, S.; Benvidi, A. Comparison of two fabricated aptasensors based on modified carbon paste/oleic acid and magnetic bar carbon paste/Fe3O4@ oleic acid nanoparticle electrodes for tetracycline detection. Biosens. Bioelectron. 2016, 85, 553–562. [Google Scholar] [CrossRef]
- Pimenta, A.M.; Souto, M.R.S.; Catarino, R.I.; Leal, M.F.C.; Lima, J.L.C. Ofloxacin determination in urine, serum and pharmaceuticals using an automatic flow potentiometric system. Anal. Sci. 2013, 29, 893–898. [Google Scholar] [CrossRef] [Green Version]
- Ismail, F.; Adeloju, S.B. Comparison of Single Layer and Bilayer Biosensors Based on Crosslinking of Penicillinase for Potentiometric Detection of Penicillin in Milk and Antibiotics. Electroanalysis 2015, 27, 1523–1531. [Google Scholar] [CrossRef]
- Almeida, S.A.; Truta, L.A.; Queirós, R.B.; Montenegro, M.; Cunha, A.L.; Sales, M.G.F. Optimizing potentiometric ionophore and electrode design for environmental on-site control of antibiotic drugs: Application to sulfamethoxazole. Biosens. Bioelectron. 2012, 35, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Regatos, D. Biosensores Ópticos de Alta Sensibilidad Basados en Técnicas de Modulación Plasmónica. Ph.D. Thesis, Universidad de Santiago de Compostela, Barcelona, Spain, 2012. [Google Scholar]
- Kivirand, K.; Floren, A.; Kagan, M.; Avarmaa, T.; Rinken, T.; Jaaniso, R. Analyzing the biosensor signal in flows: Studies with glucose optrodes. Talanta 2015, 131, 74–80. [Google Scholar] [CrossRef]
- Damborsky, P.; Svitel, J.; Katrlik, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Liu, W.; Wang, Y.; Tang, J.; Shen, G.; Yu, R. Optical fiber sensor for tetracycline antibiotics based on fluorescence quenching of covalently immobilized anthracene. Analyst 1998, 123, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Popp, J.; Pletz, M.W.; Frosch, T. Highly sensitive broadband Raman sensing of antibiotics in step-index hollow-core photonic crystal fibers. ACS Photonics 2017, 4, 138–145. [Google Scholar] [CrossRef]
- Hao, X.-J.; Zhou, X.-H.; Zhang, Y.; Long, F.; Song, L.; Shi, H.-C. Portable and reusable optofluidics-based biosensing platform for ultrasensitive detection of sulfadimidine in dairy products. Sensors 2015, 15, 8302–8313. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Myszka, D.G. Advances in surface plasmon resonance biosensor analysis. Curr. Opin. Biotechnol. 2000, 11, 54–61. [Google Scholar] [CrossRef]
- Cappi, G.; Spiga, F.M.; Moncada, Y.; Ferretti, A.; Beyeler, M.; Bianchessi, M.; Decosterd, L.; Buclin, T.; Guiducci, C. Label-free detection of tobramycin in serum by transmission-localized surface plasmon resonance. Anal. Chem. 2015, 87, 5278–5285. [Google Scholar] [CrossRef]
- De-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.; Tuñón-Blanco, P. SPR sensing of small molecules with modified RNA aptamers: Detection of neomycin B. Biosens. Bioelectron. 2009, 24, 2547–2553. [Google Scholar] [CrossRef]
- Faalnouri, S.; Cimen, D.; Bereli, N.; Denizli, A. Surface Plasmon Resonance Nanosensors for Detecting Amoxicillin in Milk Samples with Amoxicillin Imprinted Poly(hydroxyethyl methacrylate-N-methacryloyl-(L)- glutamic acid). ChemistrySelect 2020, 5, 4761–4769. [Google Scholar] [CrossRef]
- Dumont, V.; Huet, A.-C.; Traynor, I.; Elliott, C.; Delahaut, P. A surface plasmon resonance biosensor assay for the simultaneous determination of thiamphenicol, florefenicol, florefenicol amine and chloramphenicol residues in shrimps. Anal. Chim. Acta 2006, 567, 179–183. [Google Scholar] [CrossRef]
- Verma, R.; Gupta, B.D. Optical fiber sensor for the detection of tetracycline using surface plasmon resonance and molecular imprinting. Analyst 2013, 138, 7254–7263. [Google Scholar] [CrossRef]
- Shrivastav, A.M.; Usha, S.P.; Gupta, B.D. Highly sensitive and selective erythromycin nanosensor employing fiber optic SPR/ERY imprinted nanostructure: Application in milk and honey. Biosens. Bioelectron. 2017, 90, 516–524. [Google Scholar] [CrossRef]
- Pathak, A.; Parveen, S.; Gupta, B.D. Fibre optic SPR sensor using functionalized CNTs for the detection of SMX: Comparison with enzymatic approach. Plasmonics 2018, 13, 189–202. [Google Scholar] [CrossRef]
- Sharifi, M.; Dolatabadi, J.E.N.; Fathi, F.; Zakariazadeh, M.; Barzegar, A.; Rashidi, M.; Tajalli, H.; Rashidi, M.-R. Surface plasmon resonance and molecular docking studies of bovine serum albumin interaction with neomycin: Kinetic and thermodynamic analysis. Bioimpacts BI 2017, 7, 91. [Google Scholar] [CrossRef] [Green Version]
- Markina, N.E.; Markin, A.V.; Weber, K.; Popp, J.; Cialla-May, D. Liquid-liquid extraction-assisted SERS-based determination of sulfamethoxazole in spiked human urine. Anal. Chim. Acta 2020, 1109, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V.J.A. Sample pretreatment and SERS-based detection of ceftriaxone in urine. Anal. Bioanal. Chem. 2018, 410, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Rong, M.; Zhang, H.; Chen, N.; Pang, F.; Chen, Z.; Wang, T.; Yan, J. In vivo Raman measurement of levofloxacin lactate in blood using a nanoparticle-coated optical fiber probe. Biomed. Opt. Express 2016, 7, 810–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Feng, S.; Chen, Z.; Li-Chan, E.C.; Grant, E.; Lu, X. Detection and quantification of chloramphenicol in milk and honey using molecularly imprinted polymers: Canadian penny-based SERS nano-biosensor. J. Food Sci. 2014, 79, N2542–N2549. [Google Scholar] [CrossRef]
- El-Zahry, M.R.; Refaat, I.H.; Mohamed, H.A.; Rosenberg, E.; Lendl, B. Utility of surface enhanced Raman spectroscopy (SERS) for elucidation and simultaneous determination of some penicillins and penicilloic acid using hydroxylamine silver nanoparticles. Talanta 2015, 144, 710–716. [Google Scholar] [CrossRef]
- Qian, J.; Xing, C.; Ge, Y.; Li, R.; Li, A.; Yan, W. Gold nanostars-enhanced Raman fingerprint strip for rapid detection of trace tetracycline in water samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 232, 118146. [Google Scholar] [CrossRef]
- Alwan, A.M.; Wali, L.A.; Hasan, K.K. A new route for developing highly efficient nano biochemical sensors for detecting ultra-low concentrations of tetracycline antibiotic residue in water. Gold Bull. 2020, 53, 39–46. [Google Scholar] [CrossRef]
- Wang, W.; Sang, Q.; Yang, M.; Du, J.; Yang, L.; Jiang, X.; Han, X.; Zhao, B. Detection of several quinolone antibiotic residues in water based on Ag-TiO2 SERS strategy. Sci. Total Environ. 2020, 702, 134956. [Google Scholar] [CrossRef]
- Karaseva, N.; Ermolaeva, T.; Mizaikoff, B. Piezoelectric sensors using molecularly imprinted nanospheres for the detection of antibiotics. Sens. Actuators B Chem. 2016, 225, 199–208. [Google Scholar] [CrossRef]
- Karaseva, N.A.; Belyaeva, E.A.; Levkina, V.V.; Soboleva, I.G.; Ermolaeva, T.N. Development of Piezoelectric Sensors on the Basis of Electrosynthesized Molecularly Imprinted Polymers for β-lactam Antibiotics’ Detection. Procedia Technol. 2017, 27, 185–186. [Google Scholar] [CrossRef]
- Ebarvia, B.S.; Ubando, I.E. Molecularly Imprinted Polymer Sensing Layer for Tetracycline Chemical Sensor Based on Piezoelectric Quartz Crystal Transducer. Sens. Transducers 2018, 28, 7–11. [Google Scholar]
- Gfeller, K.Y.; Nugaeva, N.; Hegner, M. Rapid biosensor for detection of antibiotic-selective growth of Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 2626–2631. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Datar, R.H.; Hansen, K.M.; Thundat, T.; Cote, R.J.; Majumdar, A. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat. Biotechnol. 2001, 19, 856–860. [Google Scholar] [CrossRef]
- Duffy, J. Nanomechanical Label Free Micro RNA Detection for Cancer and Liver Injury Diagnosis. Ph.D. Thesis, Trinity College Dublin, Dublin, Ireland, 2018. [Google Scholar]
- Kosaka, P.M.; Calleja, M.; Tamayo, J. Optomechanical devices for deep plasma cancer proteomics. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2018; pp. 26–38. [Google Scholar]
- Shiwarski, D.J.; Tashman, J.W.; Tsamis, A.; Bliley, J.M.; Blundon, M.A.; Aranda-Michel, E.; Jallerat, Q.; Szymanski, J.M.; McCartney, B.M.; Feinberg, A.W. yFibronectin-Based Nanomechanical Biosensors to Map 3D Strains in Live Cells and Tissues. BioRxiv 2020. [Google Scholar] [CrossRef]
- Chen, X.; Pan, Y.; Liu, H.; Bai, X.; Wang, N.; Zhang, B. Label-free detection of liver cancer cells by aptamer-based microcantilever biosensor. Biosens. Bioelectron. 2016, 79, 353–358. [Google Scholar] [CrossRef]


| Antibiotic | Resistance Genes | Mechanism of Resistance | Bacteria | Ref |
|---|---|---|---|---|
| Aminoglycosides | ||||
| Gentamicin, neomycin, kanamycin, tobramycin, amikacin | aac (6′) - Ie + aph (2″)-Ia | Encodes a bifunctional enzyme, AAC(6′)-APH(2”), that confers resistance to a broad spectrum of aminoglycosides, able to acetylate and phosphorylate antibiotics | Staphylococcus aureus Enterococcus spp., Staphylococcus aureus, Streptococcus agalactiae (group B), Streptococcus mitis and group G Streptococcus | [122,123] |
| Neomycin, kanamycin, tobramycin, amikacin | ant(4′)-Ia | Plasmid-encoded aminoglycoside nucleotidyltransferase | S. aureus, S. epidermidis, S. aureus, Enterococcus spp. and Bacillus spp. | [124] |
| Neomycin, kanamycin | aph(3′)-IIIa | Aminoglycoside 3′-phosphotransferase | S. aureus, Enterococcus faecalis | [125] |
| Amikacin | aac (6′) - Ie + aph (3″) | Modification of the amikacin molecule by acyltransferases | Acinetobacter baumannii | [126] |
| Glucopeptides | ||||
| Vancomycin, teicoplanin | vanA | Encodes a D-alanine-D-alanine ligase of modified specificity that synthesizes peptidoglycan precursors with reduced affinity for glycopeptide antibiotics | Enterococcus faecium, Enterococcus faecalis, S. aureus | [127] |
| Vancomycin | vanB | Synthesis of modified peptidoglycan precursors terminating in D-lactate. | Enterococcus faecalis V583 | [128] |
| vanC | Synthesize a dipeptide which is incorporated into peptidoglycan precursors | Enterococcus gallinarum | [129] | |
| vanE | Synthesis of modified peptidoglycan precursors | Enterococcus faecalis | [130] | |
| vanD | Synthesized peptidoglycan precursors terminating in D-lactate. | Enterococcus faecium BM4339 | [131] | |
| vanG | The inducible synthesis of peptidoglycan precursors ending in D-alanine-D-serine | Enterococcus faecalis BM4518 and WCH9 | [132] | |
| vanM | Encodes a D-alanine-D-alanine ligase of modified specificity that synthesizes peptidoglycan precursors with reduced affinity for glycopeptide antibiotics | Enterococcus faecium | [133] | |
| vanL | D-Ala-D-Ser, which is incorporated into peptidoglycan precursors, which subsequently have a low binding affinity for vancomycin | Enterococcus faecalis N06-0364 | [134] | |
| Βeta-lactams | ||||
| Carbapenem | bla OXA-23 | Production of carbapenemases | Acinetobacter baumannii | [135] |
| blaOXA-24 | [136] | |||
| blaOXA-51 | [137] | |||
| blaOXA-58 | Carbapenem-hydrolyzing oxacillinase | [138] | ||
| bla KPC | Production of carbapenemases | Klebsiella pneumoniae, Enterobacteriaceae | [139,140] | |
| mexR | Multidrug resistance operon repressor MexR | Pseudomonas aeruginosa | [141,142] | |
| Quinolones | ||||
| Fluoroquinolones | gyrA, gyrB, parC, and parE | Alteration in enzymes, alterations in efflux pump activity | Salmonella, K. Pneumoniae, A. Baumannii, P. Aeruginosa, Neisseria gonorrhoeae, Escherichia coli | [143,144,145] |
| qnr | Encodes a pentapeptide repeat protein that protects DNA gyrase from inhibition by fluoroquinolones | K. pneumoniae, Enterococcus faecalis | [146,147] | |
| Oxazolidinone | ||||
| Linezolid | cfr | Methyltransferase activity | Staphylococcus aureus, Enterococcus spp. | [148,149] |
| Lipopeptide | ||||
| Daptomycin | mprF | Increase in the lysyl-phosphatidyl glycerol production | Staphylococcus aureus | [150] |
| yyG (walk) | Synthesis of a histidine kinase sensor | |||
| rpoB and rpoC | Shown to cause cell-wall thickening and reduction in the negative charge of the outer layer | |||
| cls2 | Encodes for a cardiolipin synthase | |||
| agrA | Encodes a quorum sensing system | |||
| pgsA | Synthesis of phosphatidyl glycerol | |||
| pnpA | Encodes for a polynucleotide phosphorylase | |||
| dltABCD | Involved in cell-wall teichoic acid D-alanination | |||
| cls, gdpD | Encoding enzymes of phospholipid metabolism | Enterococcus | [151] | |
| Polymyxin | ||||
| Colistin | arnBCADTEF operon and pmrE | Modification of the lipid A with aminoarabinose | Salmonella enterica, Klebsiella pneumoniae, Escherichia coli, Proteus mirabilis, Proteeae bacteria, Serratia marcescens and P. aeruginosa | [41,152,153,154,155] |
| pmrAB, pmrD, phoPQ, parRS, mcr | L-Ara4N and PEtn modification of lipid A | E. coli, Salmonella enterica, P. aeruginosa | [41,156] | |
| Dlt-ABCD, graXSR, dra/dlt, liaSR, and CiaR operons | Adding D-alanine (D-Ala) to teichoic acids, thereby increasing net positive charge | Staphylococcus aureus, Bordetella pertussis, Streptococcus gordonii, Listeria monocytogenes and Group B Streptococcus | [41,157] | |
| siaD, cps operon, ompA, kpnEF, phoPQ, and rcs | Loss of polymyxin target and capsule polysaccharide (CPS) overproduction | Neisseria meningitidis, K. pneumoniae and S. enterica | [41,158] | |
| spgM, pgm, hldA, hldD, oprH, cj1136, waaF, lgtF, galT, cstII, galU | Lipooligosaccharide (LOS) and LPS modification | Salmonella typhimurium, Campylobacter jejuni and Haemophilus influenzae | [41,158,159] | |
| Antibiotic | Adverse Effects | Dose | Cmax | Ref |
|---|---|---|---|---|
| Aminoglycosides | ||||
| Gentamicin | Nephrotoxicity, Neurotoxicity, Ototoxicity | 5–7 mg/kg/day | 5–10 mg/L | [201,202] |
| Amikacin | 15–20 mg/kg/day | 20–35 mg/L | [201,203] | |
| Tobramycin | 5–7 mg/kg/day | 5–10 mg/L | [201,204] | |
| Glycopeptides | ||||
| Vancomycin | Nephrotoxicity, Ototoxicity, Severe vesicular reactions, Hemorrhagic occlusive retinal vasculitis | 15–20 mg/kg/12 h | 20–50 mg/L | [170,171,205,206] |
| Teicoplanin | Nephrotoxicity, Ototoxicity, Thrombocytopenia | 43 mg/L | [205,207,208] | |
| Polymyxins | ||||
| Colistin | Nephrotoxicity, Neurotoxicity | 150mg (single dose) | 18 µg/mL | [209] |
| β-Lactamics | ||||
| Penicillins | ||||
| Ampicillin-sulbactam | Thrombocytopenia, eosinophilia, leukopenia, and transient elevation of transaminases | 1000:500 mg | 8–37 µg/mL | [210] |
| Cephalosporins | ||||
| Cephalexin | At high doses, coagulation disorders, platelet function disorders, leukopenias, thrombocytopenias, neutropenias, decreased hemoglobin and hematocrit, hemolytic anemias. Nephrotoxicity | 0.25 g/6 h | 14 µg/mL | [211,212,213,214] |
| Cephradine | 0.5–2g/6 h | 12 µg/mL | ||
| Cefoxitin | 1–2 g/6–8 h | 20 µg/mL | ||
| Cefuroxime | 0.5–1g/6–8 h | 40 µg/mL | ||
| Ceftazidime | 1–2 g/8–12 h | 120 µg/mL | ||
| Moxalactam | 500–200 mg/kg//6–12 hr | 100 µg/mL | ||
| Carbapenems | ||||
| Imipenem | In high doses, neurological toxicity, seizures rarely occur. Hematological alterations, such as leukopenia, eosinophilia, or thrombocytosis, moderate and transient increases in transaminases, alkaline phosphatase. Doripenem is toxic by epidermal necrolysis and Steven-Johnson syndrome | 1 g | 69.9 mg/L | [215,216] |
| Meropenem | 1 g | 61.6 mg/L | ||
| Ertapenem | 1 g | 164.6 mg/L | ||
| Doripenem | 500 mg | 23 mg/L | ||
| Quinolones | ||||
| Pipemidic acid | In some cases, tendinitis or tendon rupture. Fatal ventricular arrhythmias and neurotoxicity infrequently. Some quinolones that cause problems of phototoxicity (clinafloxacin), liver (trovafloxacin), or cardiac (grapafloxacin) toxicity have been withdrawn from the market | 400 mg | 4 mg/L | [217,218,219,220,221,222] |
| Ciprofloxacin | 400 mg | 1.6 mg/L | ||
| Ofloxacin | 400 mg | 4 mg/L | ||
| Levofloxacin | 500 mg | 5 mg/L | ||
| Oxazolidinone | ||||
| Linezolid | Hematological toxicity, mitochondrial toxicity in blood cells and nerve fibers of the skin, hypoglycemia, lactic acidosis, and acute pancreatitis | 1.5 mg/Kg | 2.5 mg/L | [223,224,225,226] |
| Lipopeptide | ||||
| Daptomycin | Muscle toxicity. Neurological disorders (paraesthesia, dysesthesia) and eosinophilic pneumonia, skin and subcutaneous tissue disorders, hepatobiliary disorders, musculoskeletal, and connective tissue disorders. | 4 mg/kg/day | 62.4 µg/mL | [227] |
| Type of Biosensor | Antibiotic | Biosensor Characteristics | Matrix | Limit/Detection Range | Ref |
|---|---|---|---|---|---|
| Electrochemical | Aminoglycosides | RNA aptamers | Blood | 2-6 µM | [241] |
| Penicillin G | Gold NP, catalytic hydrolysis | Buffer | 4.5nM | [242] | |
| Chloramphenicol and Kanamycin | Antibodies as bioreceptors | Buffer | 45 pg/mL and 6.31 pg/mL | [24] | |
| Amikacin | Copper oxide modified carbon paste electrode | Buffer | 1 µM | [243] | |
| Fluoroquinolones | Antibody modified magnetic beads | Milk | 0.009 µg/L | [244] | |
| Tobramycin | RNA aptamers | Human serum | 0.7 µM | [245] | |
| Ciprofloxacin | Antibodies on a poly (pyrrole-NHS) film | Buffer | 10 pg/mL | [246] | |
| Penicillin | Capture—SELEX (DNA aptamers) | Milk | 0.17 µg/L | [247] | |
| Magnetic graphene gold NP | Milk | 0.057 ng/mL | [248] | ||
| Tetracyclines | Carbon and oleic acid electrode antibodies | Milk | 3.8 fM | [249] | |
| Ofloxacin | Automatic flow potentiometric system | Urine and serum | 1 μM | [250] | |
| Optical | Tetracycline Oxytetracycline Doxycycline | Fiber optic biosensor. Copolymer containing anthracene | Commercial samples and urine | 1 μM 2 μM 2 μM | [251] |
| Doxorubicin Daunorubicin | Fluorescence-induced LED fiber optic | Buffer | 18 ng/mL 13 ng/mL | [23] | |
| Moxifloxacin | Hollow core photonic crystal fiber optic | Aqueous solution | 682.43 ng/mL | [252] | |
| Vancomycin | Molecular imprinted polymer NP functionalized fiber optic grids (LPFG—MIP NPs) | Blood plasma | 0.0032 ng/mL | [22] | |
| Sulfadimidine | Portable and reusable optofluidic-based biosensor platform | Dairy products | 0.05 ng/L | [253] | |
| Tobramycin | Portable resonance plasmon setup (T-LSPR) coupled to DNA aptamers | Patient serum | 3.4 µM | [254] | |
| Amoxicillin | SPR. Polymeric film (hydroxyethyl methacrylate-N-methacryloyl-(L) -glutamic acid) | Milk | 0.0012 ng/mL | [255] | |
| Erythromycin | Fiber optic SPR/ERY printed nanostructure | Milk Honey | 47.41 ng/mL 28.48 ng/mL | [256] | |
| SERS | Sulfamethoxazole | Hydroxylamine stabilized silver NP | Enriched urine | 1.7 µg/mL | [257] |
| Ceftriaxone | Gold NP | Urine | 0.7µM | [258] | |
| Ampicillin Penicillin G Carbenicillin Penicilloic acid | Hydroxylamine and silver NP | Deionized water | 27 ng/mL 29 ng/mL 30 ng/mL 28 ng/mL | [259] | |
| Tetracycline | Raman fingerprint strip coated with anti-tetracycline mAb | Water | 0.04 ng/mL | [260] | |
| Macroporous silicon and gold NP | Water | 1 nM | [261] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garzón, V.; Bustos, R.-H.; G. Pinacho, D. Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring. J. Pers. Med. 2020, 10, 147. https://doi.org/10.3390/jpm10040147
Garzón V, Bustos R-H, G. Pinacho D. Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring. Journal of Personalized Medicine. 2020; 10(4):147. https://doi.org/10.3390/jpm10040147
Chicago/Turabian StyleGarzón, Vivian, Rosa-Helena Bustos, and Daniel G. Pinacho. 2020. "Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring" Journal of Personalized Medicine 10, no. 4: 147. https://doi.org/10.3390/jpm10040147
APA StyleGarzón, V., Bustos, R. -H., & G. Pinacho, D. (2020). Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring. Journal of Personalized Medicine, 10(4), 147. https://doi.org/10.3390/jpm10040147