Laboratory-Based Resources for COVID-19 Diagnostics: Traditional Tools and Novel Technologies. A Perspective of Personalized Medicine
Abstract
:1. Introduction
2. Laboratory Testing as a Basis for the Diagnosis, Treatment, and Monitoring of COVID-19
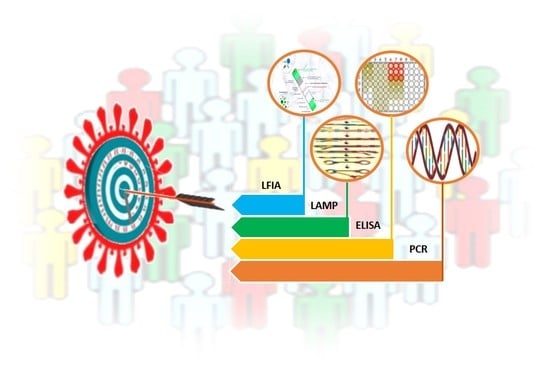
3. Laboratory-Based Tests to Diagnose COVID-19
4. Molecular Technologies for Identification of Nucleic Acids
5. Serological Methods for Detecting Antibodies and Determining Protective Immunity in SARS-CoV-2 Infected Patients
6. Search and Development of New Methods for Rapidly Diagnosing COVID-19
7. A Year of Fighting the COVID-19 Pandemic: Is It Already Time for Personalized Medicine?
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO) Official Website. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200701-covid-19-sitrep-163.pdf?sfvrsn=c202f05b2 (accessed on 2 September 2020).
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. 2019 Novel Coronavirus, Wuhan, China. Information for Healthcare Professionals. Available online: https://www.cdc.gov/coronavirus/2019-nCoV/hcp/index.html (accessed on 11 September 2020).
- European Centre for Disease Prevention and Control (ECDC) COVID 19. Available online: https://www.ecdc.europa.eu/en/novel-coronavirus-china (accessed on 11 August 2020).
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Adriano, A.; Berhane, S.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; Beese, S.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020, 8, CD013705. [Google Scholar] [CrossRef]
- Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; Lee, T.-F.; Chung, M.-Y.; Huang, C.-H.; Chen, S.-Y.; Hsueh, P.-R.; Chen, S.-C. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. In Reference Module in Life Sciences; Elsevier: Oxford, UK, 2020. [Google Scholar] [CrossRef]
- Ghebreyesus, T.A.; Swaminathan, S. Scientists are sprinting to outpace the novel coronavirus. Lancet 2020, 395, 762–764. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Calisher, C.; Carroll, D.; Colwell, R.; Corley, R.B.; Daszak, P.; Drosten, C.; Enjuanes, L.; Farrar, J.; Field, H.; Golding, J.; et al. Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. Lancet 2020, 395, e42–e43. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.D.; Hinrichs, A.S.; Clawson, H.; Gonzalez, J.N.; Lee, B.T.; Nassar, L.R.; Raney, B.J.; Rosenbloom, K.R.; Nerli, S.; Rao, A.A.; et al. The UCSC SARS-CoV-2 Genome Browser. Nat. Genet. 2020, 52, 1–8. [Google Scholar] [CrossRef]
- Hoffman, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V.J. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.-J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeKosky, B.J. A molecular trap against COVID-19. Science 2020, 369, 1167–1168. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef]
- Noh, J.Y.; Yoon, S.W.; Kim, D.J.; Lee, M.S.; Kim, J.H.; Na, W.; Song, D.; Jeong, D.G.; Kim, H.K. Simultaneous detection of severe acute respiratory syndrome, Middle East respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch. Virol. 2017, 162, 1617–1623. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Wang, P.; Wang, X.; Geng, C.; Chen, J.; Gong, Y. Molecular diagnosis of COVID-19: Current situation and trend in China (Review). Exp. Ther. Med. 2020, 20, 1. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, M.; Shen, C.; Wang, F.; Yuan, J.; Li, J.; Zhang, M.; Wang, Z.; Xing, L.; Wei, J.; et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef] [PubMed]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Elalamy, I.; Kastritis, E.; Sergentanis, T.N.; Politou, M.; Psaltopoulou, T.; Gerotziafas, G.; Dimopoulos, M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020, 95, 834–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.H.; Best, N.; McAuley, J.; Porter, J.L.; Seemann, T.; Schultz, M.B.; Sait, M.; Orlando, N.; Mercoulia, K.; Ballard, S.A.; et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J. Med. Microbiol. 2020, 69, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Zhou, Y.; Ye, J.; Al-Maskri, A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.K.; Pan, Y.; Cheng, S.M.; Hui, K.P.; Krishnan, P.; Liu, Y.; Ng, D.Y.; Wan, C.K.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef] [Green Version]
- Wan, Z.; Zhang, Y.N.; He, Z.; Liu, J.; Lan, K.; Hu, Y.; Zhang, C. A Melting Curve-Based Multiplex RT-qPCR Assay for Simultaneous Detection of Four Human Coronaviruses. Int. J. Mol. Sci. 2016, 17, 1880. [Google Scholar] [CrossRef] [Green Version]
- Adams, N.M.; Leelawong, M.; Benton, A.; Quinn, C.; Haselton, F.R.; Schmitz, J.E. COVID-19 diagnostics for resource-limited settings: Evaluation of “unextracted” qRT-PCR. J. Med. Virol. 2021, 93, 559–563. [Google Scholar] [CrossRef]
- Hung, D.L.-L.; Li, X.; Chiu, K.H.-Y.; Yip, C.C.-Y.; To, K.K.-W.; Chan, J.F.-W.; Sridhar, S.; Chung, T.W.-H.; Lung, K.-C.; Liu, R.W.-T.; et al. Early-Morning vs Spot Posterior Oropharyngeal Saliva for Diagnosis of SARS-CoV-2 Infection: Implication of Timing of Specimen Collection for Community-Wide Screening. Open Forum Infect. Dis. 2020, 7, ofaa210. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Med. Chem. Enzym. Inhib. 2020, 35, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on 29 December 2020).
- Alcoba-Florez, J.; Gil-Campesino, H.; de Artola, D.G.; González-Montelongo, R.; Valenzuela-Fernández, A.; Ciuffreda, L.; Flores, C. Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int. J. Infect. Dis. 2020, 99, 190–192. [Google Scholar] [CrossRef]
- Brukner, I.; Eintracht, S.; Papadakis, A.I.; Faucher, D.; Lamontagne, B.; Spatz, A.; Oughton, M. Maximizing confidence in a negative result: Quantitative sample adequacy control. J. Infect. Public Health 2020, 13, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Kelly-Cirino, C.D.; Nkengasong, J.; Kettler, H.; Tongio, I.; Gay-Andrieu, F.; Escadafal, C.; Piot, P.; Peeling, R.W.; Gadde, R.; Boehme, C. Importance of diagnostics in epidemic and pandemic preparedness. BMJ Glob. Health 2019, 4 (Suppl. 2). [Google Scholar] [CrossRef]
- Petruzzi, G.; De Virgilio, A.; Pichi, B.; Mazzola, F.; Zocchi, J.; Mercante, G.; Spriano, G.; Pellini, R. COVID -19: Nasal and oropharyngeal swab. Head Neck 2020, 42, 1303–1304. [Google Scholar] [CrossRef]
- World Health Organization, Official Website. Diagnostic Testing for SARS-CoV-2. Available online: https://apps.who.int/iris/bitstream/handle/10665/334254/WHO-2019-nCoV-laboratory-2020.6.pdf (accessed on 30 December 2020).
- Shen, Z.; Xiao, Y.; Kang, L.; Ma, W.; Shi, L.; Zhang, L.; Zhou, Z.; Yang, J.; Zhong, J.; Yang, D. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 713–720. [Google Scholar] [CrossRef] [Green Version]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Pujadas, E.; Chaudhry, F.; McBride, R.; Richter, F.; Zhao, S.; Wajnberg, A.; Nadkarni, G.; Glicksberg, B.S.; Houldsworth, J.; Cordon-Cardo, C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med. 2020, 8, e70. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Geoghegan, J.L.; Ren, X.; Storey, M.; Hadfield, J.; Jelley, L.; Jefferies, S.; Sherwood, J.; Paine, S.; Huang, S.; Douglas, J.; et al. Genomic epidemiology reveals transmission patterns and dynamics of SARS-CoV-2 in Aotearoa New Zealand. Nat. Commun. 2020, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.F.; Tseng, S.P.; Yen, C.H.; Yang, J.Y.; Tsao, C.H.; Shen, C.W.; Chen, K.H.; Liu, F.T.; Liu, W.T.; Chen, Y.M.; et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014, 451, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Isabel, M.; Damien, G.; Benoit, K.; Hafid, D.; Soleimani, R.; Vincenzo, C.; Olivier, V.; Beatrice, G.; Fleur, W.; Hector, R.V. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J. Clin. Virol. 2020, 128, 104413. [Google Scholar]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C.; Rawlings, S.; Smith, D.; Das, S.; Strich, J.R.; Chertow, D.S.; Davey, R.T.; Cohen, J.I. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients With Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Nicol, T.; Lefeuvre, C.; Serri, O.; Pivert, A.; Joubaud, F.; Dubée, V.; Kouatchet, A.; Ducancelle, A.; Lunel-Fabiani, F.; Le Guillou-Guillemette, H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays and one rapid lateral flow immunoassay (NG Biotech). J. Clin. Virol. 2020, 129, 104511. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Monteil, V.M.; Maurer-Stroh, S.; Yew, C.W.; Leong, C.; Mohd-Ismail, N.K.; Arularasu, S.C.; Chow, V.T.K.; Lin, R.T.P.; Mirazimi, A.; et al. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2. Eurosurveillance 2020, 25, 2000291. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.N.; Chokkalingam, N.; Reuschel, E.L.; Purwar, M.; Xu, Z.; Gary, E.N.; Kim, K.Y.; Helble, M.; Schultheis, K.; Walters, J.; et al. SARS-CoV-2 Assays To Detect Functional Antibody Responses That Block ACE2 Recognition in Vaccinated Animals and Infected Patients. J. Clin. Microbiol. 2020, 58, e01533-20. [Google Scholar] [CrossRef] [PubMed]
- Harvala, H.; Robb, M.L.; Watkins, N.; Ijaz, S.; Dicks, S.; Patel, M.; Supasa, P.; Wanwisa, D.; Liu, C.; Mongkolsapaya, J.; et al. Convalescent plasma therapy for the treatment of patients with COVID -19: Assessment of methods available for antibody detection and their correlation with neutralising antibody levels. Transfus. Med. 2020. [Google Scholar] [CrossRef]
- Patel, R.; Babady, E.; Theel, E.S.; Storch, G.A.; Pinsky, B.A.; George, K.S.; Smith, T.C.; Bertuzzi, S. Report from the American Society for Microbiology COVID-19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS-CoV-2/COVID-19. mBio 2020, 11, e00722-20. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef]
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.C.; Rattigan, S.M.; Borgert, B.A.; Moreno, C.A.; Solomon, B.D.; Rodriguez-Barraquer, I.; et al. A systematic review of antibody mediated immunity to coronaviruses: Antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. MedRxiv 2020. [Google Scholar] [CrossRef]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling early humoral response to diagnose novel Coronavirus disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randolph, H.E.; Barreiro, L.B. Herd Immunity: Understanding COVID-19. Immunity 2020, 52, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Yano, T.; Semba, S.; Akachi, S.; Kobayashi, T.; Nishinaka, T.; Notomi, T.; Matsuyama, S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 2014, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Wang, H.; Jin, H.; Feng, N.; Zheng, X.; Cao, Z.; Li, L.; Wang, J.; Yan, F.; Wang, L.; et al. Visual detection of Ebola virus using reverse transcription loop-mediated isothermal amplification combined with nucleic acid strip detection. Arch. Virol. 2016, 161, 1125–1133. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef]
- Yu, L.; Wu, S.; Hao, X.; Dong, X.; Mao, L.; Pelechano, V.; Chen, W.; Yin, X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP). Diagn. Platf. Clin. Chem. 2020, 66, 975–977. [Google Scholar] [CrossRef]
- Du, W.; Lv, M.; Li, J.; Yu, R.; Jiang, J.-H. A ligation-based loop-mediated isothermal amplification (ligation-LAMP) strategy for highly selective microRNA detection. Chem. Commun. 2016, 52, 12721–12724. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Duan, X.; Huang, J.; Huang, L.; Zhang, L.; Cheng, W.; Ding, S.; Min, X. Detection of KRAS mutation via ligation-initiated LAMP reaction. Sci. Rep. 2019, 9, 5955. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 556, eabc7075. [Google Scholar] [CrossRef] [PubMed]
- Seasun Biomaterials Inc. Official Website. Available online: http://www.seasunbio.com (accessed on 2 September 2020).
- Color Genomics, Inc. Official Website. Available online: https://www.color.com (accessed on 6 September 2020).
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.Y.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Wen, T.; Huang, C.; Shi, F.-J.; Zeng, X.-Y.; Lu, T.; Ding, S.-N.; Jiao, Y. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef]
- Andryukov, B.G. Six decades of lateral flow immunoassay: From determining metabolic markers to diagnosing COVID-19. AIMS Microbiol. 2020, 6, 280–304. [Google Scholar] [CrossRef]
- Oliveira, B.A.; Oliveira, L.C.; Sabino, E.C.; Okay, T.S. SARS-CoV-2 and the COVID-19 disease: A mini review on diagnostic methods. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e44. [Google Scholar] [CrossRef]
- Lu, H.; Stratton, C.W.; Tang, Y.-W. An evolving approach to the laboratory assessment of COVID-19. J. Med. Virol. 2020, 92, 1812–1817. [Google Scholar] [CrossRef]
- Johns Hopkins University Health Safety Center Official Website. Available online: https://www.centerforhealthsecurity.org/resources/COVID-19/serology/Serology-based-tests-for-COVID-19.html (accessed on 14 June 2020).
- Cellex Ltd. Official Website. Cellex qSARS-CoV-2 IgG/IgM Rapid Test. Available online: https://cellexcovid.com (accessed on 23 August 2020).
- ChemBio Ltd. Official Website. DPP® COVID-19 IgM/IgG System. Available online: http://chembio.com (accessed on 22 August 2020).
- Hardy Diagnostics Official Website. Anti-SARS-CoV-2 Rapid Test. Available online: https://hardydiagnostics.com/sars-cov-2 (accessed on 23 August 2020).
- Healgen Scientific LLC Official Website. COVID-19 Antibody Rapid Detection Kit. Available online: https://www.healgen.com/if-respiratory-covid-19 (accessed on 25 August 2020).
- Hangzhou Biotest Biotech Co., Ltd. Official Website. Right Sign™ COVID-19 IgM/IgG Rapid Test Kit. Available online: https://www.healgen.com/if-respiratory-covid-19 (accessed on 25 August 2020).
- Aytu Biosciences/Orient Gene Biotech Official Website. The COVID-19 IgG/IgM Point-Of-Care Rapid Test. Available online: https://stocknewsnow.com/companynews/5035338834942348/AYTU/101843 (accessed on 24 August 2020).
- Avacta Group plc Official Website. SARS-CoV-2 Rapid Antigen Test Clinical Collaboration. Available online: https://avacta.com//sars-cov-2-rapid-antigen-test-clinical-collaboration/ (accessed on 12 September 2020).
- Ogawa, J.; Zhu, W.; Tonnu, N.; Singer, O.; Hunter, T.; Ryan, A.L.; Pao, G.M. The D614G mutation in the SARS-CoV2 Spike protein increases infectivity in an ACE2 receptor dependent manner. bioRxiv 2020. [Google Scholar] [CrossRef]
- Burtscher, J.; Cappellano, G.; Omori, A.; Koshiba, T.; Millet, G.P. Mitochondria: In the Cross Fire of SARS-CoV-2 and Immunity. iScience 2020, 23, 101631. [Google Scholar] [CrossRef] [PubMed]
- Žitnik, I.P.; Černe, D.; Mancini, I.; Simi, L.; Pazzagli, M.; Di Resta, C.; Podgornik, H.; Lampret, B.R.; Podkrajšek, K.T.; Sipeky, C.; et al. Personalized laboratory medicine: A patient-centered future approach. Clin. Chem. Lab. Med. 2018, 56, 1981–1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, S.O.; Van Hal, S.J. Personalized Medicine and Infectious Disease Management. Trends Microbiol. 2017, 25, 875–876. [Google Scholar] [CrossRef] [PubMed]
- Al-Mozaini, M.A.; Mansour, M.K. Personalized medicine. Is it time for infectious diseases? Saudi Med. J. 2016, 37, 1309–1311. [Google Scholar] [CrossRef]
- Florindo, H.F.; Kleiner, R.; Vaskovich-Koubi, D.; Acúrcio, R.C.; Carreira, B.; Yeini, E.; Tiram, G.; Liubomirski, Y.; Satchi-Fainaro, R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020, 15, 630–645. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Battagello, D.S.; Dragunas, G.; Klein, M.O.; Ayub, A.L.; Velloso, F.J.; Correa, R.G. Unpuzzling COVID-19: Tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission. Clin. Sci. 2020, 134, 2137–2160. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef]
- Alshahawey, M.; Raslan, M.; Sabri, N. Sex-mediated effects of ACE2 and TMPRSS2 on the incidence and severity of COVID-19; The need for genetic implementation. Curr. Res. Transl. Med. 2020, 68, 149–150. [Google Scholar] [CrossRef]
- Luo, H.; Zhao, M.; Tan, D.; Liu, C.; Yang, L.; Qiu, L.; Gao, Y.; Yu, H. Anti-COVID-19 drug screening: Frontier concepts and core technologies. Chin. Med. 2020, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zeberg, H.; Pääbo, S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nat. Cell Biol. 2020, 587, 610–612. [Google Scholar] [CrossRef] [PubMed]
- Anastassopoulou, C.; Gkizarioti, Z.; Patrinos, G.P.; Tsakris, A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genom. 2020, 14, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Delanghe, J.R.; Speeckaert, M.M.; De Buyzere, M.L. ACE Ins/Del genetic polymorphism and epidemiological findings in COVID-19. Clin. Chem. Lab. Med. 2020, 58, 1129–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ma, Q.; Han, H.; Su, H.; Liu, F.; Wu, K.; Wang, W.; Zhu, C. The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clin. Chem. Lab. Med. 2020, 58, 1121–1124. [Google Scholar] [CrossRef] [Green Version]
- Kočar, E.; Režen, T.; Rozman, D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 158849. [Google Scholar] [CrossRef]
- Wendt, R.; Kalbitz, S.; Lübbert, C.; Kellner, N.; Macholz, M.; Schroth, S.; Ermisch, J.; Latosisnka, A.; Arnold, B.; Mischak, H.; et al. Urinary Proteomics Associates with COVID-19 Severity: Pilot Proof-of-Principle Data and Design of a Multicentric Diagnostic Study. Proteomics 2020, 20, e2000202. [Google Scholar] [CrossRef]
- Wu, K.; Zou, J.; Chang, H.Y. RNA-GPS Predicts SARS-CoV-2 RNA Localization to Host Mitochondria and Nucleolus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef]
- Maggi, E.; Canonica, G.W.; Moretta, L. COVID-19: Unanswered questions on immune response and pathogenesis. J. Allergy Clin. Immunol. 2020, 146, 18–22. [Google Scholar] [CrossRef]
- Blot, M.; LYMPHONIE Study Group; Bour, J.-B.; Quenot, J.P.; Bourredjem, A.; Nguyen, M.; Guy, J.; Monier, S.; Georges, M.; Large, A.; et al. The dysregulated innate immune response in severe COVID-19 pneumonia that could drive poorer outcome. J. Transl. Med. 2020, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Olwenyi, O.A.; Dyavar, S.R.; Acharya, A.; Podany, A.T.; Fletcher, C.V.; Ng, C.L.; Reid, S.P.; Byrareddy, S.N. Immuno-epidemiology and pathophysiology of coronavirus disease 2019 (COVID-19). J. Mol. Med. 2020, 98, 1–15. [Google Scholar] [CrossRef] [PubMed]




| Classes of Antibodies against SARS-CoV-2 | Estimated Level of Seroconversion | Mean Time of Seroconversion (Days) | Seroconversion within 15 Days after Symptom Onset |
|---|---|---|---|
| Total antibodies | 93.1% (161/173) | 11 | 100.0% |
| IgM | 82.7% (143/173) | 12 | 94.3% |
| IgG | 64.7% (112/173) | 15 | 79.8% |
| Criteria | LAMP | PCR |
|---|---|---|
| Temperature cycles | Isothermal amplification (60–65 °С) | Different temperature cycles required |
| Number of primers | 4(6) specially designed primers | 2 primers |
| Analysis time | Up to 45 min | Up to 6–8 h |
| DNA output | DNA yield—10–20 μg | DNA yield—up to 0.2 μg |
| Visual detection | Possible | Impossible |
| Economy and ease of use | Economical and easy to carry out | Requires expensive equipment and trained personnel |
| Sample inhibitor sensitivity | Insensitive | Sensitive |
| Multiplexing capability | Possible | Impossible |
| Knowledge of the method, clinical evaluation | Little known, clinical evaluation ongoing | Well known, clinically proven |
| Country of Origin and Developer Company | Name | Description | Refs. |
|---|---|---|---|
| Melbourne University, Australia | N1-STOP-LAMP | Qualitative (RT-LAMP) detection of the CDC N1 region of the nucleocapsid (N) SARS-CoV-2 gene in oral and nasopharyngeal mucus samples, infection. Result <20 min. | [70] |
| Seasun Biomaterials Inc., South Korea | AQ-TOP™ COVID-19 Rapid Detection Kit. | Qualitative (RT-LAMP) detection of RNA in samples of mucus from oropharynx and nasopharynx, BAL fluid and sputum in the acute phase of infection. Result <30 min. | [71] |
| Color Genomics, Inc., USА | Color Genomics SARS-COV-2 RT-LAMP Diagnostic Assay | Qualitative (RT-LAMP) detection of RNA in samples of mucus from oropharynx and nasopharynx, bronchoalveolar lavage (BAL) fluid and sputum in the acute phase of infection. Result <30 min. | [72] |
| Country of Origin and Developer Company | Sensitivity/Specificity of Test Systems (%) | Description | Used Biosubstrates, Analysis Time | Refs. |
|---|---|---|---|---|
| US/China, Cellex Inc. | 93.8/95.6 | Detection of IgM/IgG to the nucleocapsid SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, Na citrate), 20 min | [79] |
| US, ChemBio | 92.7 (IgM) и 95.9 (IgG)/99.0 (IgM и IgG) | Detection of IgM/IgG to the nucleocapsid SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, heparin), 15 min | [80] |
| US, Autobio Diagnostics Co. Ltd. (+ Hardy Diagnostics) | 95.7 (IgM) и 99.0 (IgG)/99.0 (IgM и IgG) | Detection of IgM/IgG к antigenic proteins SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, heparin), 15 min | [81] |
| US/China, Healgen Scientific LLC | 96.7 (IgG), 86.7 (IgM), 96.7/98.0 (IgG), 99.0 (IgM), 97.0 | Detection of IgM/IgG к antigenic proteins SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, heparin), 10 min | [82] |
| China, Hangzhou Biotest Biotech Co., Ltd. | 92.5 (IgM), 91.56 (IgG)/98.1 (IgM), 99.52 (IgG) | Detection of IgM/IgG to S1 locus of S protein SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, heparin), 20 min | [83] |
| US/China, Aytu Biosciences/Orient Gene Biotech | 87.9 (IgM) & 97.2 (IgG)/100.0 (IgG и IgM | Detection of IgM/IgG к antigenic proteins SARS-CoV-2 | Serum, plasma, whole blood (K2-EDTA, Na citrate), 10 min | [84] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andryukov, B.G.; Besednova, N.N.; Kuznetsova, T.A.; Fedyanina, L.N. Laboratory-Based Resources for COVID-19 Diagnostics: Traditional Tools and Novel Technologies. A Perspective of Personalized Medicine. J. Pers. Med. 2021, 11, 42. https://doi.org/10.3390/jpm11010042
Andryukov BG, Besednova NN, Kuznetsova TA, Fedyanina LN. Laboratory-Based Resources for COVID-19 Diagnostics: Traditional Tools and Novel Technologies. A Perspective of Personalized Medicine. Journal of Personalized Medicine. 2021; 11(1):42. https://doi.org/10.3390/jpm11010042
Chicago/Turabian StyleAndryukov, Boris G., Natalya N. Besednova, Tatyana A. Kuznetsova, and Ludmila N. Fedyanina. 2021. "Laboratory-Based Resources for COVID-19 Diagnostics: Traditional Tools and Novel Technologies. A Perspective of Personalized Medicine" Journal of Personalized Medicine 11, no. 1: 42. https://doi.org/10.3390/jpm11010042
APA StyleAndryukov, B. G., Besednova, N. N., Kuznetsova, T. A., & Fedyanina, L. N. (2021). Laboratory-Based Resources for COVID-19 Diagnostics: Traditional Tools and Novel Technologies. A Perspective of Personalized Medicine. Journal of Personalized Medicine, 11(1), 42. https://doi.org/10.3390/jpm11010042