Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18
Abstract
:1. Introduction
2. Results
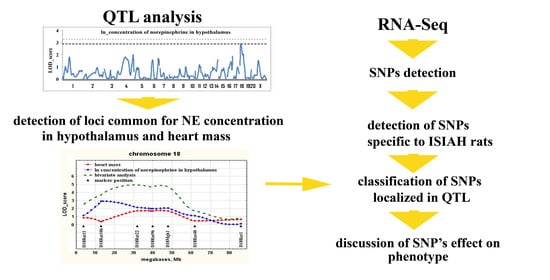
2.1. Determination of Loci Associated with the Concentration of Norepinephrine in the Hypothalamus
2.2. Determination of Loci Associated with both the Concentration of Norepinephrine in the Hypothalamus and Heart Mass
2.3. Nucleotide Substitutions (SNPs) Detected in ISIAH but not in WAG Rats
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Determination of Norepinephrine Concentration in the Hypothalamus
4.3. QTL (Quantitative Trait Locus) Analysis
4.4. Tissue Collection for SNP Analysis
4.5. RNA Sequencing
4.6. Polymorphism Detection
4.7. Prediction of the SNP Effects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morgan, H.E.; Baker, K.M. Cardiac hypertrophy. Mechanical, neural, and endocrine dependence. Circulation 1991, 83, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, S.; Tarazi, R.C.; Khairallah, P.A.; Bumpus, F.M. Cardiac hypertrophy in spontaneously hypertensive rats. Circ. Res. 1974, 35, 775–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamide, K.; Rakugi, H.; Higaki, J.; Okamura, A.; Nagai, M.; Moriguchi, K.; Ohishi, M.; Satoh, N.; Tuck, M.L.; Ogihara, T. The renin-angiotensin and adrenergic nervous system in cardiac hypertrophy in fructose-fed rats. Am. J. Hypertens. 2002, 15, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Simpson, P.C.; Kariya, K.; Karns, L.R.; Long, C.S.; Karliner, J.S. Adrenergic hormones and control of cardiac myocyte growth. Mol. Cell. Biochem. 1991, 104, 35–43. [Google Scholar] [CrossRef] [PubMed]
- DeLalio, L.J.; Sved, A.F.; Stocker, S.D. Sympathetic Nervous System Contributions to Hypertension: Updates and Therapeutic Relevance. Can. J. Cardiol. 2020, 36, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Macefield, V.G.; Henderson, L.A. Identification of the human sympathetic connectome involved in blood pressure regulation. Neuroimage 2019, 202, 116119. [Google Scholar] [CrossRef]
- Bear, M.H.; Reddy, V.; Bollu, P.C. Neuroanatomy, Hypothalamus; StatPearls Publishing LLC: Treasure Island, FL, USA, 2020. [Google Scholar]
- Atzori, M.; Cuevas-Olguin, R.; Esquivel-Rendon, E.; Garcia-Oscos, F.; Salgado-Delgado, R.C.; Saderi, N.; Miranda-Morales, M.; Trevino, M.; Pineda, J.C.; Salgado, H. Locus Ceruleus Norepinephrine Release: A Central Regulator of CNS Spatio-Temporal Activation? Front. Synaptic. Neurosci. 2016, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Pacak, K.; Palkovits, M.; Kopin, I.J.; Goldstein, D.S. Stress-induced norepinephrine release in the hypothalamic paraventricular nucleus and pituitary-adrenocortical and sympathoadrenal activity: In vivo microdialysis studies. Front. Neuroendocrinol. 1995, 16, 89–150. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, C.; Weiss, G.L.; Fu, X.; Fisher, M.O.; Begley, J.C.; Stevens, C.R.; Harrison, L.M.; Tasker, J.G. Acute stress desensitizes hypothalamic CRH neurons to norepinephrine and physiological stress. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Selvetella, G.; Lembo, G. Mechanisms of cardiac hypertrophy. Heart Fail. Clin. 2005, 1, 263–273. [Google Scholar] [CrossRef]
- Morris, M.; Ross, J.; Sundberg, D.K. Catecholamine biosynthesis and vasopressin and oxytocin secretion in the spontaneously hypertensive rat: An in vitro study of localized brain regions. Peptides 1985, 6, 949–955. [Google Scholar] [CrossRef]
- Oparil, S.; Chen, Y.F.; Peng, N.; Wyss, J.M. Anterior hypothalamic norepinephrine, atrial natriuretic peptide, and hypertension. Front. Neuroendocrinol. 1996, 17, 212–246. [Google Scholar] [CrossRef] [PubMed]
- McCrossan, Z.A.; Billeter, R.; White, E. Transmural changes in size, contractile and electrical properties of SHR left ventricular myocytes during compensated hypertrophy. Cardiovasc. Res. 2004, 63, 283–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacak, K.; Yadid, G.; Jakab, G.; Lenders, J.W.; Kopin, I.J.; Goldstein, D.S. In vivo hypothalamic release and synthesis of catecholamines in spontaneously hypertensive rats. Hypertension 1993, 22, 467–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, N.D.; Mukherjee, K.; Ganguly, P.K. Norepinephrine levels in paraventricular nucleus of spontaneously hypertensive rats: Role of neuropeptide Y. Am. J. Physiol. 1993, 265, H893–H898. [Google Scholar] [CrossRef]
- Arabia, A.M.; Catapano, L.; Storini, C.; Perego, C.; De Luigi, A.; Head, G.A.; De Simoni, M.G. Impaired central stress-induced release of noradrenaline in rats with heart failure: A microdialysis study. Neuroscience 2002, 114, 591–599. [Google Scholar] [CrossRef]
- Buzueva, I.I.; Filyushina, E.E.; Shmerling, M.D.; Markel, A.L.; Yakobson, G.S. Chromogranin location in the adrenal glands of ISIAH rats. Bull. Exp. Biol. Med. 2013, 154, 393–395. [Google Scholar] [CrossRef]
- Bilek, R.; Safarik, L.; Ciprova, V.; Vlcek, P.; Lisa, L. Chromogranin A, a member of neuroendocrine secretory proteins as a selective marker for laboratory diagnosis of pheochromocytoma. Physiol Res. 2008, 57 (Suppl. 1), S171–S179. [Google Scholar]
- Markel, A.L.; Redina, O.E.; Gilinsky, M.A.; Dymshits, G.M.; Kalashnikova, E.V.; Khvorostova, Y.V.; Fedoseeva, L.A.; Jacobson, G.S. Neuroendocrine profiling in inherited stress-induced arterial hypertension rat strain with stress-sensitive arterial hypertension. J. Endocrinol. 2007, 195, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Suslonova, O.V.; Roshchevskaya, I.M.; Rasputina, A.A. Morphometry of the heart ventricles in NISAG rats during the early postnatal ontogenesis. Isvestiia Komi Nauchnogo Cent. URO RAN 2016, 1, 45–50. [Google Scholar]
- Shmerling, M.D.; Buzueva, I.I.; Korostyshevskaia, I.M.; Lazarev, V.A.; Maksimov, V.F.; Filiushina, E.E.; Markel, A.L.; Iakobson, G.S. Stereomorphometric study of target organs in rats with hereditary stress-induced arterial hypertension at different periods of postnatal ontogenesis under changed conditions of nursing. Morfologiia 2005, 128, 85–90. [Google Scholar] [PubMed]
- Esler, M. The sympathetic system and hypertension. Am. J. Hypertens 2000, 13, 99S–105S. [Google Scholar] [CrossRef] [Green Version]
- Ryazanova, M.; Klimov, L.; Seryapina, A.; Zarytova, V.; Repkova, M.; Levina, A.; Markel, A. Nanocomposite (Si-ODN-II), containing antisense oligonucleotides targeted to mRNA of the beta-1-adrenoreceptors for hypertension therapy in rats with stress-sensitive arterial hypertension (ISIAH rats). J. Hypertens. 2018, 36 (Suppl. 3), e81–e82. [Google Scholar] [CrossRef]
- Redina, O.E.; Machanova, N.A.; Efimov, V.M.; Markel, A.L. Rats with inherited stress-induced arterial hypertension (ISIAH strain) display specific quantitative trait loci for blood pressure and for body and kidney weight on chromosome 1. Clin. Exp. Pharmacol. Physiol. 2006, 33, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Redina, O.E.; Smolenskaya, S.E.; Maslova, L.N.; Markel, A.L. The genetic control of blood pressure and body composition in rats with stress-sensitive hypertension. Clin. Exp. Hypertens. 2013, 35, 484–495. [Google Scholar] [CrossRef]
- Redina, O.E.; Smolenskaya, S.E.; Maslova, L.N.; Markel, A.L. Genetic Control of the Corticosterone Level at Rest and Under Emotional Stress in ISIAH Rats with Inherited Stress-Induced Arterial Hypertension. Clin. Exp. Hypertens. 2010, 32, 364–371. [Google Scholar] [CrossRef]
- Redina, O.E.; Smolenskaya, S.E.; Jacobson, G.S.; Markel, A.L. Norepinephrine content in the hypothalamus and medulla oblongata of ISIAH rats is regulated by several genetic loci. Bull. Exp. Biol. Med. 2009, 148, 223–226. [Google Scholar] [CrossRef]
- Ershov, N.I.; Markel, A.L.; Redina, O.E. Strain-Specific Single-Nucleotide Polymorphisms in Hypertensive ISIAH Rats. Biochemistry 2017, 82, 224–235. [Google Scholar] [CrossRef]
- Duong, C.; Charron, S.; Xiao, C.; Hamet, P.; Ménard, A.; Roy, J.; Deng, A.Y. Distinct quantitative trait loci for kidney, cardiac, and aortic mass dissociated from and associated with blood pressure in Dahl congenic rats. Mamm Genome 2006, 17, 1147–1161. [Google Scholar] [CrossRef]
- Hirooka, Y. Sympathetic Activation in Hypertension: Importance of the Central Nervous System. Am. J. Hypertens. 2020, 33, 914–926. [Google Scholar] [CrossRef]
- Stoll, M.; Cowley, A.W.J.; Tonellato, P.J.; Greene, A.S.; Kaldunski, M.L.; Roman, R.J.; Dumas, P.; Schork, N.J.; Wang, Z.; Jacob, H.J. A genomic-systems biology map for cardiovascular function. Science 2001, 294, 1723–1726. [Google Scholar] [CrossRef] [PubMed]
- Charron, S.; Lambert, R.; Eliopoulos, V.; Duong, C.; Menard, A.; Roy, J.; Deng, A.Y. A loss of genome buffering capacity of Dahl salt-sensitive model to modulate blood pressure as a cause of hypertension. Hum. Mol. Genet. 2005, 14, 3877–3884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, A.Y.; Nattel, S.; Shi, Y.; L’Heureux, N.; Cardin, S.; Menard, A.; Roy, J.; Tardif, J.C. Distinct genomic replacements from Lewis correct diastolic dysfunction, attenuate hypertension, and reduce left ventricular hypertrophy in Dahl salt-sensitive rats. J. Hypertens. 2008, 26, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, P.; Voigt, B.; Kloting, I. Novel quantitative trait loci for blood pressure and related traits on rat chromosomes 1, 10, and 18. Biochem. Biophys. Res. Commun. 1997, 235, 343–348. [Google Scholar] [CrossRef]
- Fisher, J.P.; Paton, J.F. The sympathetic nervous system and blood pressure in humans: Implications for hypertension. J. Hum. Hypertens. 2012, 26, 463–475. [Google Scholar] [CrossRef] [Green Version]
- Gurgerian, S.V.; Vatinian, S. The multifactorial genesis of left ventricle remodeling in patients with essential arterial hypertension. Kardiologiia 2013, 53, 38–42. [Google Scholar]
- Cohen, J.D.; Egan, B.M. The role of sympathetic activation in cardiovascular disease. Postgrad. Med. 2003, 114, 4–10. [Google Scholar] [CrossRef]
- Saxena, A.; Moshynska, O.; Sankaran, K.; Viswanathan, S.; Sheridan, D.P. Association of a novel single nucleotide polymorphism, G(-248)A, in the 5’-UTR of BAX gene in chronic lymphocytic leukemia with disease progression and treatment resistance. Cancer Lett. 2002, 187, 199–205. [Google Scholar] [CrossRef]
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. 3’ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J. 2008, 27, 482–498. [Google Scholar] [CrossRef] [Green Version]
- Greenbaum, L.; Smith, R.C.; Rigbi, A.; Strous, R.; Teltsh, O.; Kanyas, K.; Korner, M.; Lancet, D.; Ben-Asher, E.; Lerer, B. Further evidence for association of the RGS2 gene with antipsychotic-induced parkinsonism: Protective role of a functional polymorphism in the 3’-untranslated region. Pharm. J. 2009, 9, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Mendelova, A.; Holubekova, V.; Grendar, M.; Zubor, P.; Svecova, I.; Loderer, D.; Snahnicanova, Z.; Biringer, K.; Danko, J.; Lasabova, Z. Association between 3’UTR polymorphisms in genes ACVR2A, AGTR1 and RGS2 and preeclampsia. Gen. Physiol. Biophys. 2018, 37, 185–192. [Google Scholar] [CrossRef]
- Han, Y.J.; Ma, S.F.; Wade, M.S.; Flores, C.; Garcia, J.G. An intronic MYLK variant associated with inflammatory lung disease regulates promoter activity of the smooth muscle myosin light chain kinase isoform. J. Mol. Med. 2012, 90, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.J.; Lee, S.Y.; Choi, J.E.; Kang, H.G.; Do, S.K.; Lee, J.H.; Yoo, S.S.; Lee, E.B.; Seok, Y.; Cho, S.; et al. Intronic variant of EGFR is associated with GBAS expression and survival outcome of early-stage non-small cell lung cancer. Thorac. Cancer 2018, 9, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006, 7, 61–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Fuscoe, J.C.; Zhao, C.; Guo, C.; Jia, M.; Qing, T.; Bannon, D.I.; Lancashire, L.; Bao, W.; Du, T.; et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014, 5, 3230. [Google Scholar] [CrossRef] [PubMed]
- Valdez, B.C.; Henning, D.; So, R.B.; Dixon, J.; Dixon, M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 2004, 101, 10709–10714. [Google Scholar] [CrossRef] [Green Version]
- Chauvet, C.; Menard, A.; Deng, A.Y. Two candidate genes for two quantitative trait loci epistatically attenuate hypertension in a novel pathway. J. Hypertens. 2015, 33, 1791–1801. [Google Scholar] [CrossRef]
- Jacob, H.J.; Lindpaintner, K.; Lincoln, S.E.; Kusumi, K.; Bunker, R.K.; Mao, Y.P.; Ganten, D.; Dzau, V.J.; Lander, E.S. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell 1991, 67, 213–224. [Google Scholar] [CrossRef]
- Brown, D.M.; Provoost, A.P.; Daly, M.J.; Lander, E.S.; Jacob, H.J. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat. Genet. 1996, 12, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Yagil, C.; Sapojnikov, M.; Kreutz, R.; Katni, G.; Lindpaintner, K.; Ganten, D.; Yagil, Y. Salt susceptibility maps to chromosomes 1 and 17 with sex specificity in the Sabra rat model of hypertension. Hypertension 1998, 31, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Chauvet, C.; Crespo, K.; Menard, A.; Roy, J.; Deng, A.Y. Modularization and epistatic hierarchy determine homeostatic actions of multiple blood pressure quantitative trait loci. Hum. Mol. Genet. 2013, 22, 4451–4459. [Google Scholar] [CrossRef] [Green Version]
- Te Pas, M.F.; Madsen, O.; Calus, M.P.; Smits, M.A. The Importance of Endophenotypes to Evaluate the Relationship between Genotype and External Phenotype. Int. J. Mol. Sci. 2017, 18, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergquist, J.; Sciubisz, A.; Kaczor, A.; Silberring, J. Catecholamines and methods for their identification and quantitation in biological tissues and fluids. J. Neurosci. Methods 2002, 113, 1–13. [Google Scholar] [CrossRef]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Basten, C.J.; Weir, B.S.; Zeng, Z.-B. QTL Cartographer, Version 1.17; Department of Statistics, North. Carolina State University: Raleigh, NC, USA, 2004. [Google Scholar]
- Churchill, G.A.; Doerge, R.W. Empirical Threshold Values for Quantitative Trait Mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef]
- Lander, E.; Kruglyak, L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat. Genet. 1995, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Mather, K.; Jinks, J.L. Introduction to Biometrical Genetics; Chapman & Hall: London, UK, 1977; p. 231. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, R.A.; Weinstock, G.M.; Metzker, M.L.; Muzny, D.M.; Sodergren, E.J.; Scherer, S.; Scott, G.; Steffen, D.; Worley, K.C.; Burch, P.E.; et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004, 428, 493–521. [Google Scholar] [CrossRef] [Green Version]
- Hermsen, R.; de Ligt, J.; Spee, W.; Blokzijl, F.; Schafer, S.; Adami, E.; Boymans, S.; Flink, S.; van Boxtel, R.; van der Weide, R.H.; et al. Genomic landscape of rat strain and substrain variation. BMC Genom. 2015, 16, 357. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]



| Trait | ISIAH M ± SEM (n = 10) | WAG M ± SEM (n = 14) |
|---|---|---|
| Concentration of norepinephrine in the hypothalamus, ng/mg of tissue | 1.698 ± 0.068 ** | 1.045 ± 0.068 |
| Chr. | Peak Marker (Mb) | Confidence Interval *, Mb | LOD Score | p | Variability, % |
|---|---|---|---|---|---|
| Significant QTL | |||||
| 18 | D18Rat106 (13.2) | 0–50 | 2.91 | 0.05 | 10.8 |
| Suggestive QTL | |||||
| 1 | D1Rat30 (107.1) | 90–128 | 1.82 | 0.05 | 6.6 |
| 4 | D4Rat27 (137.8) | 84–186 | 1.80 | 0.05 | 8.0 |
| 4 | D4Rat68 (233.3) | 214–242 | 1.99 | 0.025 | 7.2 |
| 6 | D6Rat143 (48.1) | 40–72 | 1.60 | 0.05 | 10.7 |
| 6 | D6Rat75 (136.6) | 120–156.9 | 1.71 | 0.05 | 7.8 |
| 14 | D14Rat18 (86.0) | 68–115.1 | 1.58 | 0.025 | 6.3 |
| 16 | D16Rat48 (79.7) | 72–90 | 1.87 | 0.025 | 7.3 |
| X | DXRat26 (45.1) | 28–70 | 1.54 | 0.05 | 6.1 |
| Trait Measurement in F2 Hybrids (ISIAH × WAG) n | Chr. | Peak Marker (Mb) | Genotype | D # | ||
|---|---|---|---|---|---|---|
| I/I M ± SEM n | I/W M ± SEM n | W/W M ± SEM n | ||||
| 1.93 ± 0.07 123 | Significant QTL | |||||
| 18 | D18Rat106 (13.2) | 2.21 ± 0.15 ** 29 | 1.97 ± 0.10 56 | 1.65 ± 0.11 38 | 0.1 | |
| Suggestive QTL | ||||||
| 1 | D1Rat30 (107.1) | 1.63 ± 0.12 †† 27 | 2.05 ± 0.08 71 | 1.94 ± 0.18 24 | −1.7 | |
| 4 | D4Rat27 (137.8) | 1.80 ± 0.11 * 31 | 1.79 ± 0.08 56 | 2.27 ± 0.16 †† 36 | 1.0 | |
| 4 | D4Rat68 (233.3) | 1.87 ± 0.13 * 25 | 1.80 ± 0.07 74 | 2.39 ± 0.21 †† 24 | 1.3 | |
| 6 | D6Rat143 (48.1) | 1.96 ± 0.11 * 50 | 2.01 ± 0.10 58 | 1.54 ± 0.13 †† 15 | 1.2 | |
| 6 | D6Rat75 (136.6) | 2.11 ± 0.14 † 31 | 1.75 ± 0.08 59 | 2.09 ± 0.14 † 33 | −35.0 | |
| 14 | D14Rat18 (86.0) | 1.72 ± 0.08 †† 30 | 2.14 ± 0.12 58 | 1.76 ± 0.09 † 35 | −20.0 | |
| 16 | D16Rat48 (79.7) | 1.61 ± 0.09 **†† 31 | 2.03 ± 0.10 60 | 2.06 ± 0.13 32 | −0.9 | |
| X | DXRat26 (45.1) | 2.17 ± 0.13 ** 52 | 1.75 ± 0.06 71 | |||
| Chr. | Peak Marker (Mb) | Confidence Interval *, Mb | LOD Score | p Chromosome-Wise | Variability, % |
|---|---|---|---|---|---|
| Heart Mass | |||||
| 18 | D18Mgh1 (47.7) | 18–60 | 1.76 | 0.025 | 6.5 |
| SnpEff Classification | Number of SNPs | Effect |
|---|---|---|
| 3_prime_UTR_variant | 55 | modifier |
| 5_prime_UTR_premature_start_codon_gain_variant | 3 | low |
| 5_prime_UTR_variant | 3 | modifier |
| downstream_gene_variant | 67 | modifier |
| intergenic_region | 10 | modifier |
| intron_variant | 25 | modifier |
| missense_variant | 13 | moderate |
| synonymous_variant | 58 | low |
| upstream_gene_variant | 6 | modifier |
| Gene Symbol | Position | ID | DP | SNP | Amino Acid Substitution | SIFT Score * | SIFT Classification |
|---|---|---|---|---|---|---|---|
| Slc4a9 | 29059899 | novel | 118 | c.269C>T | p.Ala90Val | 0.021 | deleterious |
| Dcp2 | 35336920 | novel | 76 | c.1109C>T | p.Ala370Val | 0.718 | tolerated |
| Megf10 | 51550880 | rs199133377 | 131 | c.3404C>G | p.Thr1135Ser | 0.823 | tolerated |
| RGD1312005 | 53356293 | rs8169475 | 634 | c.361A>G | p.Asn121Asp | 1.000 | tolerated |
| Synpo | 55104621 | rs63909326 | 331 | c.1727T>C | p.Ile576Thr | 0.378 | tolerated |
| Synpo | 55104676 | rs198024246 | 253 | c.1672C>T | p.His558Tyr | 1.000 | tolerated |
| Tcof1 | 55324879 | rs198780519 | 351 | c.2828A>G | p.Asn943Ser | 1.000 | tolerated |
| Tcof1 | 55334042 | rs197009609 | 274 | c.1556C>A | p.Ala519Glu | 0.014 | deleterious |
| Tcof1 | 55347410 | rs199120971 | 231 | c.126T>G | p.His42Gln | 1.000 | tolerated |
| Csf1r | 55682600 | rs198399348 | 798 | c.1825C>A | p.Leu609Met | 0.486 | tolerated |
| Hmgxb3 | 55690668 | rs198759766 | 350 | c.3513T>G | p.His1171Gln | 0.265 | tolerated |
| Hmgxb3 | 55715142 | rs197391293 | 293 | c.1337G>A | p.Gly446Asp | 0.087 | tolerated |
| Napg | 57562399 | rs198931177 | 570 | c.448T>A | p.Cys150Ser | 0.157 | tolerated |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redina, O.E.; Smolenskaya, S.E.; Polityko, Y.K.; Ershov, N.I.; Gilinsky, M.A.; Markel, A.L. Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18. J. Pers. Med. 2021, 11, 67. https://doi.org/10.3390/jpm11020067
Redina OE, Smolenskaya SE, Polityko YK, Ershov NI, Gilinsky MA, Markel AL. Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18. Journal of Personalized Medicine. 2021; 11(2):67. https://doi.org/10.3390/jpm11020067
Chicago/Turabian StyleRedina, Olga E., Svetlana E. Smolenskaya, Yulia K. Polityko, Nikita I. Ershov, Michael A. Gilinsky, and Arcady L. Markel. 2021. "Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18" Journal of Personalized Medicine 11, no. 2: 67. https://doi.org/10.3390/jpm11020067
APA StyleRedina, O. E., Smolenskaya, S. E., Polityko, Y. K., Ershov, N. I., Gilinsky, M. A., & Markel, A. L. (2021). Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18. Journal of Personalized Medicine, 11(2), 67. https://doi.org/10.3390/jpm11020067