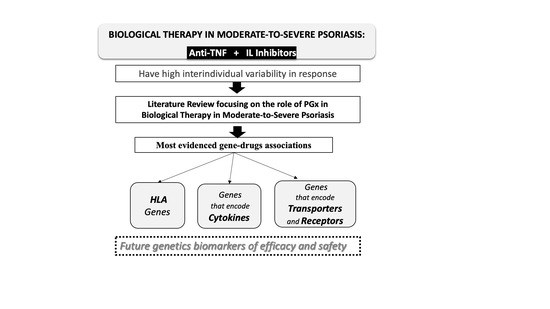
Influence of Genetic Polymorphisms on Response to Biologics in Moderate-to-Severe Psoriasis
Abstract
:1. Introduction
2. Materials and Methods
3. Pharmacogenetics of Biological Therapies in Psoriasis
3.1. Human Leukocyte Antigens (HLAs)
3.2. Cytokines and Associated Proteins
3.2.1. Tumor Necrosis Factor (TNF)
3.2.2. Interleukin 1 Beta (IL1B)
3.2.3. Interleukin 6 (IL6)
3.2.4. Interleukin 12B (IL12B)
3.2.5. Interleukin 17 (IL17) Genes
3.2.6. Tumor Necrosis Factor Alpha-Induced Protein 3 (TNFAIP3)
3.3. Transporters, Receptors and Associated Proteins
3.3.1. Phosphodiesterase 3A (PDE3A)-Solute Carrier Organic Anion Transporter Family Member 1C1 (SLCO1C1)
3.3.2. Solute Carrier Family 12 Member 8 (SLC12A8)
3.3.3. Tumor Necrosis Factor Receptor Superfamily Member 1B (TNFRSF1B)
3.3.4. CD84 Molecule (CD84)
3.3.5. Fc Fragment of IgG Receptors IIA and IIIA (FCGR2A and FCGR3A)
3.3.6. Interleukin 17 Receptor A (IL17RA)
3.3.7. Interleukin 23 Receptor (IL23R)
3.3.8. Toll-Like Receptors
Toll-Like Receptor 2 (TLR2)
Lymphocyte Antigen 96 (LY96)
TIR Domain-Containing Adapter Protein (TIRAP)
Toll-Like Receptor 5 (TLR5)
Toll-Like Receptor 9 (TLR9)
3.3.9. Peptidoglycan Recognition Protein 4 (PGLYRP4)
3.3.10. F-Box and Leucine-Rich Repeat Protein 19 (FBXL19)
3.4. Other Genes
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nast, A.; Gisondi, P.; Ormerod, A.; Saiag, P.; Smith, C.; Spuls, P.; Arenberger, P.; Bachelez, H.; Barker, J.; Dauden, E.; et al. European S3-Guidelines on the systemic treatment of psoriasis vulgaris—Update 2015—Short version—EDF in cooperation with EADV and IPC. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2277–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezzolo, E.; Naldi, L. Epidemiology of major chronic inflammatory immune-related skin diseases in 2019. Expert Rev. Clin. Immunol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Feldman, S.R. Psoriasis causes as much disability as other major medical diseases. J. Am. Acad. Dermatol. 2020, 82, 256–257. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, J.N.; De Paula, A.P.; Martins, G.A. Psoriatic arthritis in patients with psoriasis: Evaluation of clinical and epidemiological features in 133 patients followed at the University Hospital of Brasília. An. Bras. Dermatol. 2012, 87, 539–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henes, J.C.; Ziupa, E.; Eisfelder, M.; Adamczyk, A.; Knaudt, B.; Jacobs, F.; Lux, J.; Schanz, S.; Fierlbeck, G.; Spira, D.; et al. High prevalence of psoriatic arthritis in dermatological patients with psoriasis: A cross-sectional study. Rheumatol. Int. 2014, 34, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Molina-Leyva, A.; Salvador-Rodriguez, L.; Martinez-Lopez, A.; Ruiz-Carrascosa, J.C.; Arias-Santiago, S. Association between psoriasis and sexual and erectile dysfunction in epidemiologic studies: A systematic review. JAMA Dermatol. 2019, 155, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Rendon, A.; Schäkel, K. Psoriasis pathogenesis and treatment. Int. J. Mol. Sci. 2019, 20, 1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harden, J.L.; Krueger, J.G.; Bowcock, A.M. The immunogenetics of psoriasis: A comprehensive review. J. Autoimmun. 2015, 64, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Chandra, A.; Ray, A.; Senapati, S.; Chatterjee, R. Genetic and epigenetic basis of psoriasis pathogenesis. Mol. Immunol. 2015, 64, 313–323. [Google Scholar] [CrossRef]
- Caputo, V.; Strafella, C.; Termine, A.; Dattola, A.; Mazzilli, S.; Lanna, C.; Cosio, T.; Campione, E.; Novelli, G.; Giardina, E.; et al. Overview of the molecular determinants contributing to the expression of psoriasis and psoriatic arthritis phenotypes. J. Cell. Mol. Med. 2020, 24, 13554–13563. [Google Scholar] [CrossRef]
- Di Meglio, P.; Villanova, F.; Nestle, F.O. Psoriasis. Cold Spring Harb Perspect. Med. 2014, 4, a015354. [Google Scholar] [CrossRef] [Green Version]
- Morizane, S.; Yamasaki, K.; Mühleisen, B.; Kotol, P.F.; Murakami, M.; Aoyama, Y.; Iwatsuki, K.; Hata, T.; Gallo, R.L. Cathelicidin antimicrobial peptide LL-37 in psoriasis enables keratinocyte reactivity against TLR9 ligands. J. Investig. Dermatol. 2012, 132, 135–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehncke, W.-H. Etiology and pathogenesis of psoriasis. Rheum. Dis. Clin. N. Am. 2015, 41, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.D.; Bowcock, A.M. Psoriasis genetics: Breaking the barrier. Trends Genet. 2010, 26, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Alwan, W.; Nestle, F.O. Pathogenesis and treatment of psoriasis: Exploiting pathophysiological pathways for precision medicine. Clin. Exp. Rheumatol. 2015, 33, S2–S6. [Google Scholar] [PubMed]
- Atwan, A.; Piguet, V.; Finlay, A.; Francis, N.; Ingram, J.R. Dermatology Life Quality Index (DLQI) as a psoriasis referral triage tool. Br. J. Dermatol. 2017, 177, e136–e137. [Google Scholar] [CrossRef] [Green Version]
- Daudén, E.; Puig, L.; Ferrándiz, C.; Sánchez-Carazo, J.L.; Hernanz-Hermosa, J.M. Venereology SPGotSAoDa. Consensus document on the evaluation and treatment of moderate-to-severe psoriasis: Psoriasis Group of the Spanish Academy of Dermatology and Venereology. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 1–18. [Google Scholar] [CrossRef]
- Kechichian, E.; Ezzedine, K. Vitamin D and the skin: An update for dermatologists. Am. J. Clin. Dermatol. 2018, 19, 223–235. [Google Scholar] [CrossRef]
- Chiricozzi, A.; Pitocco, R.; Saraceno, R.; Nistico, S.P.; Giunta, A.; Chimenti, S. New topical treatments for psoriasis. Expert Opin. Pharmacother. 2014, 15, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Linares-Pineda, T.M.; Cañadas-Garre, M.; Sánchez-Pozo, A.; Calleja-Hernández, M.Á. Gene polymorphisms as predictors of response to biological therapies in psoriasis patients. Pharmacol. Res. 2016, 113, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Iannone, L.F.; Bennardo, L.; Palleria, C.; Roberti, R.; De Sarro, C.; Naturale, M.D.; Dastoli, S.; Donato, L.; Manti, A.; Valenti, G.; et al. Safety profile of biologic drugs for psoriasis in clinical practice: An Italian prospective pharmacovigilance study. PLoS ONE 2020, 15, e0241575. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.; No, D.J.; Egeberg, A.; Wu, J.J. Choosing first-line biologic treatment for moderate-to-severe psoriasis: What does the evidence say? Am. J. Clin. Dermatol. 2017, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wcisło-Dziadecka, D.; Grabarek, B.; Kruszniewska-Rajs, C.; Strzałka-Mrozik, B. The analysis of the therapeutic potential of ustekinumab in psoriasis vulgaris treatment. Dermatol. Ther. 2019, 32, e12843. [Google Scholar] [CrossRef]
- Bai, F.; Li, G.G.; Liu, Q.; Niu, X.; Li, R.; Ma, H. Short-term efficacy and safety of IL-17, IL-12/23, and IL-23 inhibitors brodalumab, secukinumab, ixekizumab, ustekinumab, guselkumab, tildrakizumab, and risankizumab for the treatment of moderate to severe plaque psoriasis: A systematic review and network meta-analysis of randomized controlled trials. J. Immunol. Res. 2019, 2019, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Jeon, C.; Sekhon, S.; Yan, D.; Afifi, L.; Nakamura, M.; Bhutani, T. Monoclonal antibodies inhibiting IL-12, -23, and -17 for the treatment of psoriasis. Hum. Vaccines Immunother. 2017, 13, 2247–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, L.M.; Malottki, K.; Sabry-Grant, C.; Yasmeen, N.; Wright, E.; Sohrt, A.; Borg, E.; Warren, R.B. Assessing the relative efficacy of interleukin-17 and interleukin-23 targeted treatments for moderate-to-severe plaque psoriasis: A systematic review and network meta-analysis of PASI response. PLoS ONE 2019, 14, e0220868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasquillo, O.Y.; Pabón-Cartagena, G.; Falto-Aizpurua, L.A.; Santiago-Vázquez, M.; Cancel-Artau, K.J.; Arias-Berrios, G.; Martín-García, R.F. Treatment of erythrodermic psoriasis with biologics: A systematic review. J. Am. Acad. Dermatol. 2020, 83, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.W.; Puig, L.; Joshi, A.; Skup, M.; Williams, D.; Li, J.; Betts, K.A.; Augustin, M. Comparison of biologics and oral treatments for plaque psoriasis: A meta-analysis. JAMA Dermatol. 2020, 156, 258–269. [Google Scholar] [CrossRef]
- Sbidian, E.; Chaimani, A.; Afach, S.; Doney, L.; Dressler, C.; Hua, C.; Mazaud, C.; Phan, C.; Hughes, C.; Riddle, D. Systemic pharmacological treatments for chronic plaque psoriasis: A network meta-analysis. Cochrane Database Syst. Rev. 2017, 12, CD011535. [Google Scholar] [CrossRef]
- Cui, L.; Chen, R.; Subedi, S.; Yu, Q.; Gong, Y.; Chen, Z.; Shi, Y. Efficacy and safety of biologics targeting IL-17 and IL-23 in the treatment of moderate-to-severe plaque psoriasis: A systematic review and meta-analysis of randomized controlled trials. Int. Immunopharmacol. 2018, 62, 46–58. [Google Scholar] [CrossRef]
- Ten Bergen, L.L.; Petrovic, A.; Krogh Aarebrot, A.; Appel, S. The TNF/IL-23/IL-17 axis-Head-to-head trials comparing different biologics in psoriasis treatment. Scand. J. Immunol. 2020, 92, e12946. [Google Scholar] [CrossRef]
- Sawyer, L.; Cornic, L.; Levin, L.; Gibbons, C.; Møller, A.; Jemec, G. Long-term efficacy of novel therapies in moderate-to-severe plaque psoriasis: A systematic review and network meta-analysis of PASI response. J. Eur. Acad. Dermatol. Venereol. 2018, 33, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Peleva, E.; Exton, L.S.; Kelley, K.; Kleyn, C.E.; Mason, K.J.; Smith, C.H. Risk of cancer in patients with psoriasis on biological therapies: A systematic review. Br. J. Dermatol. 2018, 178, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Daudén, E.; Carretero, G.; Rivera, R.; Ferrándiz, C.; Llamas-Velasco, M.; de la Cueva, P.; Belinchón, I.; Gómez-García, F.J.; Herrera-Acosta, E.; Ruiz-Genao, D.P.; et al. Long-term safety of nine systemic medications for psoriasis: A cohort study using the Spanish Registry of Adverse Events for Biological Therapy in Dermatological Diseases (BIOBADADERM) Registry. J. Am. Acad. Dermatol. 2020, 83, 139–150. [Google Scholar] [CrossRef]
- Wu, J.J.; Merola, J.F.; Feldman, S.R.; Menter, A.; Lebwohl, M. Treatment of psoriasis with secukinumab in challenging patient scenarios: A review of the available evidence. Dermatol. Ther. 2020, 10, 351–364. [Google Scholar] [CrossRef] [Green Version]
- Farhangian, M.E.; Feldman, S.R. Immunogenicity of biologic treatments for psoriasis: Therapeutic consequences and the potential value of concomitant methotrexate. Am. J. Clin. Dermatol. 2015, 16, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Prinz, J.C. Human leukocyte antigen-class I alleles and the autoreactive T cell response in psoriasis pathogenesis. Front. Immunol. 2018, 9, 954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, D.; CAguiar, V.R.; Bitarello, B.D.; CBrandt, D.Y.; Nunes, K. A genomic perspective on HLA evolution. Immunogenetics 2018, 70, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Ovejero-Benito, M.C.; Muñoz-Aceituno, E.; Reolid, A.; Saiz-Rodríguez, M.; Abad-Santos, F.; Daudén, E. Pharmacogenetics and pharmacogenomics in moderate-to-severe psoriasis. Am. J. Clin. Dermatol. 2018, 19, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Ovejero-Benito, M.C.; Prieto-Pérez, R.; Llamas-Velasco, M.; Belmonte, C.; Cabaleiro, T.; Román, M.; Ochoa, D.; Talegón, M.; Saiz-Rodríguez, M.; Daudén, E.; et al. Polymorphisms associated with etanercept response in moderate-to-severe plaque psoriasis. Pharmacogenomics 2017, 18, 631–638. [Google Scholar] [CrossRef]
- Chiu, H.; Huang, P.-Y.; Jee, S.-H.; Hu, C.-Y.; Chou, C.-T.; Chang, Y.-T.; Hwang, C.-Y.; Tsai, T.-F. HLA polymorphism among Chinese patients with chronic plaque psoriasis: Subgroup analysis. Br. J. Dermatol. 2012, 166, 288–297. [Google Scholar] [CrossRef]
- Prieto-Pérez, R.; Solano-López, G.; Cabaleiro, T.; Román, M.; Ochoa, D.; Talegón, M.; Baniandrés, O.; López-Estebaranz, J.L.; De La Cueva, P.; Daudén, E.; et al. New polymorphisms associated with response to anti-TNF drugs in patients with moderate-to-severe plaque psoriasis. Pharmacogenomics 2018, 18, 70–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masouri, S.; Stefanaki, I.; Ntritsos, G.; Kypreou, K.P.; Drakaki, E.; Evangelou, E.; Nicolaidou, E.; Stratigos, A.J.; Antoniou, C. A pharmacogenetic study of psoriasis risk variants in a Greek population and prediction of responses to anti-TNF-α and anti-IL-12/23 agents. Mol. Diagn. Ther. 2016, 20, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Caldarola, G.; Sgambato, A.; Fanali, C.; Moretta, G.; Farina, M.; Lucchetti, D.; Peris, K.; De Simone, C. HLA-Cw6 allele, NFkB1 and NFkBIA polymorphisms play no role in predicting response to etanercept in psoriatic patients. Pharmacogenet. Genom. 2016, 26, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.; Kelleher, J.; Fagan, M.F.; Rogers, S.; Collins, P.; Barker, J.N.W.N.; Allen, M.; Hagan, R.; Renfro, L.; Kirby, B. Genetic markers of treatment response to tumour necrosis factor-α inhibitors in the treatment of psoriasis. Clin. Exp. Dermatol. 2014, 39, 519–524. [Google Scholar] [CrossRef]
- Van Vugt, L.J.; Van den Reek, J.M.P.A.; Hannink, G.; Coenen, M.J.H.; De Jong, E.M.G.J. Association of HLA-C*06:02 status with differential response to ustekinumab in patients with psoriasis: A systematic review and meta-analysis. JAMA Dermatol. 2019, 155, 708–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, E.; Cabaleiro, T.; Roman, M.; Solano-López, G.; Abad-Santos, F.; García-Díez, A.; Daudén, E. The relationship between tumour necrosis factor (TNF)-α promoter and IL12B/IL-23R genes polymorphisms and the efficacy of anti-TNF-α therapy in psoriasis: A case-control study. Br. J. Dermatol. 2013, 169, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Coto, E.; Santos-Juanes, J.; Coto-Segura, P.; Díaz, M.; Soto, J.; Queiro, R.; Alvarez, V. Mutation analysis of the LCE3B/LCE3C genes in psoriasis. BMC Med. Genet. 2010, 11, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talamonti, M.; Botti, E.; Galluzzo, M.; Teoli, M.; Spallone, G.; Bavetta, M.; Chimenti, S.; Costanzo, A. Pharmacogenetics of psoriasis: HLA-Cw6 but not LCE3B/3C deletion nor TNFAIP3 polymorphism predisposes to clinical response to interleukin 12/23 blocker ustekinumab. Br. J. Dermatol. 2013, 169, 458–463. [Google Scholar] [CrossRef]
- Van den Reek, J.M.; Coenen, M.J.; van de L’Isle Arias, M.; Zweegers, J.; Rodijk-Olthuis, D.; Schalkwijk, J.; Vermeulen, S.H.; Joosten, I.; van de Kerkhof, P.C.; Seyger, M.M.; et al. Polymorphisms in CD84, IL12B and TNFAIP3 are associated with response to biologics in patients with psoriasis. Br. J. Dermatol. 2017, 176, 1288–1296. [Google Scholar] [CrossRef]
- Batalla, A.; Coto, E.; González-Fernández, D.; González-Lara, L.; Gómez, J.; Santos-Juanes, J.; Queiro, R.; Coto-Segura, P. The Cw6 and late-cornified envelope genotype plays a significant role in anti-tumor necrosis factor response among psoriatic patients. Pharmacogenet. Genom. 2015, 25, 313–316. [Google Scholar] [CrossRef]
- Talamonti, M.; Galluzzo, M.; Botti, E.; Pavlidis, A.; Spallone, G.; Chimenti, S.; Costanzo, A. Potential role of HLA-Cw6 in clinical response to anti-tumour necrosis factor alpha and T-cell targeting agents in psoriasis patients. Clin. Drug. Investig. 2013, 33, S71. [Google Scholar]
- Talamonti, M.; Galluzzo, M.; Zangrilli, A.; Papoutsaki, M.; Egan, C.G.; Bavetta, M.; Tambone, S.; Fargnoli, M.C.; Bianchi, L. HLA-C*06:02 does not predispose to clinical response following long-term adalimumab treatment in psoriatic patients: A retrospective cohort study. Mol. Diagn. Ther. 2017, 21, 295–301. [Google Scholar] [CrossRef]
- Talamonti, M.; Galluzzo, M.; Chimenti, S.; Costanzo, A. HLA-C*06 and response to ustekinumab in Caucasian patients with psoriasis: Outcome and long-term follow-up. J. Am. Acad. Dermatol. 2016, 74, 374–375. [Google Scholar] [CrossRef] [Green Version]
- Talamonti, M.; Galluzzo, M.; Reek, J.V.D.; De Jong, E.; Lambert, J.; Malagoli, P.; Bianchi, L.; Costanzo, A. Role of the HLA-C*06 allele in clinical response to ustekinumab: Evidence from real life in a large cohort of European patients. Br. J. Dermatol. 2017, 177, 489–496. [Google Scholar] [CrossRef]
- Chiu, H.Y.; Wang, T.S.; Chan, C.C.; Cheng, Y.P.; Lin, S.J.; Tsai, T.F. Human leucocyte antigen-Cw6 as a predictor for clinical response to ustekinumab, an interleukin-12/23 blocker, in Chinese patients with psoriasis: A retrospective analysis. Br. J. Dermatol. 2014, 171, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Huang, C.C.; Randazzo, B.; Li, S.; Szapary, P.; Curran, M.; Campbell, K.; Brodmerkel, C. HLA-C*06:02 allele and response to IL-12/23 inhibition: Results from the ustekinumab phase 3 psoriasis program. J. Invest. Dermatol. 2016, 136, 2364–2371. [Google Scholar] [CrossRef] [Green Version]
- Galluzzo, M.; Boca, A.N.; Botti, E.; Potenza, C.; Malara, G.; Malagoli, P.; Vesa, S.; Chimenti, S.; Buzoianu, A.D.; Talamonti, M.; et al. IL12B (p40) gene polymorphisms contribute to ustekinumab response prediction in psoriasis. Dermatology 2015, 232, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Dand, N.; Duckworth, M.; Baudry, D.; Russell, A.; Curtis, C.J.; Lee, S.H.; Evans, I.; Mason, K.J.; Alsharqi, A.; Becher, G.; et al. HLA-C*06:02 genotype is a predictive biomarker of biologic treatment response in psoriasis. J. Allergy Clin. Immunol. 2019, 143, 2120–2130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghi, A.; Rizzo, R.; Corazza, M.; Bertoldi, A.M.; Bortolotti, D.; Sturabotti, G.; Virgili, A.; Di Luca, D. HLA-G 14-bp polymorphism: A possible marker of systemic treatment response in psoriasis vulgaris? Preliminary results of a retrospective study. Dermatol. Ther. 2014, 27, 284–289. [Google Scholar] [CrossRef]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Boyman, O.; Hefti, H.P.; Conrad, C.; Nickoloff, B.J.; Suter, M.; Nestle, F.O. Spontaneous development of psoriasis in a new animal model shows an essential role for resident T cells and tumor necrosis factor-α. J. Exp. Med. 2004, 199, 731–736. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhou, H. A meta-analysis of the relationship between tumor necrosis factor-α polymorphisms and psoriasis. Dermatology 2021, 237, 39–45. [Google Scholar] [CrossRef]
- Song, G.G.; Seo, Y.H.; Kim, J.H.; Choi, S.J.; Ji, J.D.; Lee, Y.H. Association between TNF-α (-308 A/G, -238 A/G, -857 C/T) polymorphisms and responsiveness to TNF-α blockers in spondyloarthropathy, psoriasis and Crohn’s disease: A meta-analysis. Pharmacogenomics 2015, 16, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Gulli, R.; Spanò, F.; Lantieri, F.; Burlando, M.; Parodi, A.; Mandich, P.; Puppo, F. TNF-α gene polymorphisms: Association with disease susceptibility and response to anti-TNF-α treatment in psoriatic arthritis. J. Investig. Dermatol. 2014, 134, 2503–2509. [Google Scholar] [CrossRef] [Green Version]
- Tong, Q.; Zhao, L.; Qian, X.-D.; Zhang, L.-L.; Xu, X.; Dai, S.-M.; Cai, Q.; Zhao, D.-B. Association ofTNF-α polymorphism with prediction of response to TNF blockers in spondyloarthritis and inflammatory bowel disease: A meta-analysis. Pharmacogenomics 2013, 14, 1691–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Simone, C.; Farina, M.; Maiorino, A.; Fanali, C.; Perino, F.; Flamini, A.; Caldarola, G.; Sgambato, A. TNF-alpha gene polymorphisms can help to predict response to etanercept in psoriatic patients. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1786–1790. [Google Scholar] [CrossRef]
- Vasilopoulos, Y.; Manolika, M.; Zafiriou, E.; Sarafidou, T.; Bagiatis, V.; Krüger-Krasagaki, S.; Tosca, A.; Patsatsi, A.; Sotiriadis, D.; Mamuris, Z.; et al. Pharmacogenetic analysis of TNF, TNFRSF1A, and TNFRSF1B gene polymorphisms and prediction of response to anti-TNF therapy in psoriasis patients in the Greek population. Mol. Diagn. Ther. 2012, 16, 29–34. [Google Scholar] [CrossRef]
- Nicklin, M.J.; Barton, J.L.; Nguyen, M.; FitzGerald, M.G.; Duff, G.W.; Kornman, K. A sequence-based map of the nine genes of the human Interleukin-1 cluster. Genomics 2002, 79, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Yoshimoto, T.; Torigoe, K.; Kurimoto, M.; Matsui, K.; Hada, T.; Okamura, H.; Nakanishi, K. IL-12 synergizes with IL-18 or IL-1beta for IFN-gamma production from human T cells. Int. Immunol. 2000, 12, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Loft, N.D.; Skov, L.; Iversen, L.; Gniadecki, R.; Dam, T.N.; Brandslund, I.; Hoffmann, H.J.; Andersen, M.R.; Dessau, R.B.; Bergmann, A.C.; et al. Associations between functional polymorphisms and response to biological treatment in Danish patients with psoriasis. Pharmacogenomics 2017, 18, 494–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.P.; Lee, K.W.; Yoo, D.H.; Bae, S.C. The influence of a polymorphism at position -857 of the tumour necrosis factor gene on clinical response to etanercept therapy in rheumatoid arthritis. Rheumatology 2005, 44, 547–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Renzo, L.; Bianchi, A.; Saraceno, R.; Calabrese, V.; Cornelius, C.; Iacopino, L.; Chimenti, S.; De Lorenzo, A. -174G/C IL-6 gene promoter polymorphism predicts therapeutic response to TNF-α blockers. Pharmacogenet. Genom. 2012, 22, 134–142. [Google Scholar] [CrossRef]
- Vázquez-Vázquez, C.; Posadas-Sánchez, R.; Fragoso, J.M.; Ramírez-Bello, J.; Sánchez-Guerra, M.; Osorio-Yañez, C.; Vargas-Alarcón, G. Polymorphisms are associated with the presence of premature coronary artery disease and with cardiovascular risk factors: The genetics of atherosclerotic disease Mexican study. DNA Cell Biol. 2020, 39, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Pérez, R.; Solano-López, G.; Cabaleiro, T.; Román, M.; Ochoa, D.; Talegón, M.; Baniandrés, O.; Lopez Estebaranz, J.L.; de la Cueva, P.; Daudén, E.; et al. The polymorphism rs763780 in the IL-17F gene is associated with response to biological drugs in patients with psoriasis. Pharmacogenomics 2015, 16, 1723–1731. [Google Scholar] [CrossRef]
- Del Duca, E.; Morelli, P.; Bennardo, L.; Di Raimondo, C.; Nisticò, S.P. Cytokine pathways and investigational target therapies in Hidradenitis suppurativa. Int. J. Mol. Sci. 2020, 21, 8436. [Google Scholar] [CrossRef]
- Gu, C.; Wu, L.; Li, X. IL-17 family: Cytokines, receptors and signaling. Cytokine 2013, 64, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Hawkes, J.E.; Yan, B.Y.; Chan, T.C.; Krueger, J.G. Discovery of the IL-23/IL-17 signaling pathway and the treatment of psoriasis. J. Immunol. 2018, 201, 1605–1613. [Google Scholar] [CrossRef]
- Catanoso, M.G.; Boiardi, L.; Macchioni, P.; Garagnani, P.; Sazzini, M.; De Fanti, S.; Farnetti, E.; Casali, B.; Chiarolanza, I.; Nicoli, D.; et al. IL-23A, IL-23R, IL-17A and IL-17R polymorphisms in different psoriatic arthritis clinical manifestations in the northern Italian population. Rheumatol. Int. 2013, 33, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Batalla, A.; Coto, E.; Gómez, J.; Eirís, N.; González-Fernández, D.; Castro, C.G.-D.; Daudén, E.; Llamas-Velasco, M.; Prieto-Perez, R.; Abad-Santos, F.; et al. IL17RA gene variants and anti-TNF response among psoriasis patients. Pharmacogenomics 2018, 18, 76–80. [Google Scholar] [CrossRef]
- Batalla, A.; Coto, E.; González-Lara, L.; González-Fernández, D.; Gómez, J.; Aranguren, T.F.; Queiro, R.; Santos-Juanes, J.; López-Larrea, C.; Coto-Segura, P. Association between single nucleotide polymorphisms IL17RA rs4819554 and IL17E rs79877597 and Psoriasis in a Spanish cohort. J. Dermatol. Sci. 2015, 80, 111–115. [Google Scholar] [CrossRef]
- Hartz, P.A.; (Baltimore, MD, USA). Personal communication, 15 December 2014.
- Wertz, I.E.; O’rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef]
- Shembade, N.; Ma, A.; Harhaj, E.W. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 2010, 327, 1135–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ovejero-Benito, M.C.; Muñoz-Aceituno, E.; Reolid, A.; Fisas, L.H.; Llamas-Velasco, M.; Prieto-Pérez, R.; Abad-Santos, F.; Daudén, E. Polymorphisms associated with anti-TNF drugs response in patients with psoriasis and psoriatic arthritis. J. Eur. Acad. Dermatol. Venereol. 2019, 33, e175–e177. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.M.; Jang, E.K.; Haslam, R.J. Expression and mutagenesis of the catalytic domain of cGMP-inhibited phosphodiesterase (PDE3) cloned from human platelets. Biochem. J. 1997, 323, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Pizzagalli, F.; Hagenbuch, B.; Stieger, B.; Klenk, U.; Folkers, G.; Meier, P.J. Identification of a novel human organic anion transporting polypeptide as a high affinity thyroxine transporter. Mol. Endocrinol. 2002, 16, 2283–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julià, A.; Ferrándiz, C.; Dauden, E.; Fonseca, E.; Fernández-López, E.; Sanchez-Carazo, J.L.; Vanaclocha, F.; Puig, L.; Morenoramirez, D.; Lopez-Estebaranz, J.L.; et al. Association of the PDE3A-SLCO1C1 locus with the response to anti-TNF agents in psoriasis. Pharmacogenomics 2015, 15, 322–325. [Google Scholar] [CrossRef]
- Hewett, D.; Samuelsson, L.; Polding, J.; Enlund, F.; Smart, D.; Cantone, K.; See, C.G.; Chadha, S.; Inerot, A.; Enerback, C.; et al. Identification of a psoriasis susceptibility candidate gene by linkage disequilibrium mapping with a localized single nucleotide polymorphism map. Genomics 2002, 79, 305–314. [Google Scholar] [CrossRef]
- Cabaleiro, T.; Prietoperez, R.; Navarro, R.M.; Solano, G.; Roman, M.J.; Ochoa, D.; Abadsantos, F.; Dauden, E. Paradoxical psoriasiform reactions to anti-TNFα drugs are associated with genetic polymorphisms in patients with psoriasis. Pharmacogenomics 2016, 16, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; An, P.; Zhang, R.; He, X.; Yin, G.; Min, W. Etk/Bmx as a tumor necrosis factor receptor Type 2-specific kinase: Role in endothelial cell migration and angiogenesis. Mol. Cell. Biol. 2002, 22, 7512–7523. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, A.W.; Read, C. Pathophysiology, clinical presentation, and treatment of psoriasis: A review. JAMA 2020, 323, 1945–1960. [Google Scholar] [CrossRef] [PubMed]
- González-Lara, L.; Batalla, A.; Coto, E.; Gómez, J.; Eiris, N.; Santos-Juanes, J.; Queiro, R.; Coto-Segura, P. The TNFRSF1B rs1061622 polymorphism (p.M196R) is associated with biological drug outcome in Psoriasis patients. Arch. Dermatol. Res. 2015, 307, 405–412. [Google Scholar] [CrossRef]
- Chen, W.; Xu, H.; Wang, X.; Gu, J.; Xiong, H.; Shi, Y. The tumor necrosis factor receptor superfamily member 1B polymorphisms predict response to anti-TNF therapy in patients with autoimmune disease: A meta-analysis. Int. Immunopharmacol. 2015, 28, 146–153. [Google Scholar] [CrossRef] [PubMed]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Stahl, E.A.; Saevarsdóttir, S.; Miceli, C.; Diogo, R.; Trynka, G.; Raj, T.; Mirkov, M.U.; Canhão, H.; Ikari, K.; et al. Genome-Wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013, 9, e1003394. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, C.; Grau, S.; Jäger, C.; Sondermann, P.; Brünker, P.; Waldhauer, I.; Hennig, M.; Ruf, A.; Rufer, A.C.; Stihle, M.; et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. USA 2011, 108, 12669–12674. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Marnell, L.L.; Marjon, K.D.; Mold, C.; Du Clos, T.W.; Sun, P.D. Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature 2008, 456, 989–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Haas, M. IgG-Fc receptors and the clinical relevance of their polymorphisms. Wien Klin. Wochenschr. 2001, 113, 825–831. [Google Scholar]
- Koene, H.R.; Kleijer, M.; Algra, J.; Roos, D.; Borne, A.E.V.D.; De Haas, M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 1997, 90, 1109–1114. [Google Scholar] [CrossRef] [Green Version]
- Jiménez Morales, A.; Maldonado-Montoro, M.; Martínez de la Plata, J.E.; Pérez Ramírez, C.; Daddaoua, A.; Alarcón Payer, C.; Expósito Ruiz, M.; García Collado, C. FCGR2A/FCGR3A gene polymorphisms and clinical variables as predictors of response to tocilizumab and rituximab in patients with rheumatoid arthritis. J. Clin. Pharmacol. 2019, 59, 517–531. [Google Scholar] [CrossRef]
- Tutuncu, Z.; Kavanaugh, A.; Zvaifler, N.; Corr, M.; Deutsch, R.; Boyle, D. Fcgamma receptor type IIIA polymorphisms influence treatment outcomes in patients with inflammatory arthritis treated with tumor necrosis factor alpha-blocking agents. Arthritis Rheum. 2005, 52, 2693–2696. [Google Scholar] [CrossRef]
- Julià, M.; Guilabert, A.; Lozano, F.; Suarez-Casasús, B.; Moreno, N.; Carrascosa, J.M.; Ferrándiz, C.; Pedrosa, E.; Alsina-Gibert, M.; Mascaró, J.M. The role of Fcγ receptor polymorphisms in the response to anti–tumor necrosis factor therapy in psoriasis A pharmacogenetic study. JAMA Dermatol. 2013, 149, 1033–1039. [Google Scholar] [CrossRef]
- Batalla, A.; Coto, E.; Coto-Segura, P. Influence of Fcγ receptor polymorphisms on response to anti–tumor necrosis factor treatment in psoriasis. JAMA Dermatol. 2015, 151, 1376–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendrinou, E.; Patsatsi, A.; Zafiriou, E.; Papadopoulou, D.; Aggelou, L.; Sarri, C.; Mamuris, Z.; Kyriakou, A.; Sotiriadis, D.; Roussaki-Schulze, A.; et al. FCGR3A-V158F polymorphism is a dis-ease-specific pharmacogenetic marker for the treatment of psoriasis with Fc-containing TNFα inhibitors. Pharmacogenomics 2017, 17, 237–241. [Google Scholar] [CrossRef]
- Nirula, A.; Nilsen, J.; Klekotka, P.; Kricorian, G.; Erondu, N.; Towne, J.E.; Russell, C.B.; Martin, D.A.; Budelsky, A.L. Effect of IL-17 receptor A blockade with brodalumab in inflammatory diseases. Rheumatology 2016, 55, ii43–ii55. [Google Scholar] [CrossRef] [Green Version]
- Pușcaș, A.D.; Cătană, A.; Pușcaș, C.; Roman, I.I.; Vornicescu, C.; Șomlea, M.; Orăsan, R.I. Psoriasis: Association of interleukin-17 gene pol-ymorphisms with severity and response to treatment. Exp. Ther. Med. 2019, 18, 875–880. [Google Scholar] [PubMed] [Green Version]
- Dattola, A.; Silvestri, M.; Tamburi, F.; Amoruso, G.F.; Bennardo, L.; Nisticò, S.P. Emerging role of anti-IL23 in the treatment of psoriasis: When humanized is very promising. Dermatol. Ther. 2020, 33, e14504. [Google Scholar] [CrossRef]
- Parham, C.; Chirica, M.; Timans, J.; Vaisberg, E.; Travis, M.; Cheung, J.; Pflanz, S.; Zhang, R.; Singh, K.P.; Vega, F.; et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002, 168, 5699–5708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Vugt, L.J.; Van den Reek, J.M.P.A.; Coenen, M.J.H.; de Jong, E.M.G.J. A systematic review of pharmacogenetic studies on the response to biologics in patients with psoriasis. Br. J. Dermatol. 2018, 178, 86–94. [Google Scholar] [CrossRef]
- Lancioni, C.L.; Li, Q.; Thomas, J.J.; Ding, X.; Thiel, B.; Drage, M.G.; Pecora, N.D.; Ziady, A.G.; Shank, S.; Harding, C.V.; et al. Mycobacterium tuberculosis lipoproteins directly regulate human memory CD4+T cell activation via toll-like receptors 1 and 2. Infect. Immun. 2010, 79, 663–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziarski, R.; Wang, Q.; Miyake, K.; Kirschning, C.J.; Gupta, D. MD-2 enables Toll-like receptor 2 (TLR2)-mediated responses to lip-opolysaccharide and enhances TLR2-mediated responses to Gram-positive and Gram-negative bacteria and their cell wall components. J. Immunol. 2001, 166, 1938–1944. [Google Scholar] [CrossRef] [Green Version]
- Khor, C.C.; Chapman, S.J.; Vannberg, F.O.; Dunne, A.; Murphy, C.; Ling, E.Y.; Frodsham, A.J.; Walley, A.J.; Kyrieleis, O.; Khan, A.; et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat. Genet. 2007, 39, 523–528. [Google Scholar] [CrossRef]
- Horng, T.; Barton, G.M.; Medzhitov, R. TIRAP: An adapter molecule in the Toll signaling pathway. Nat. Immunol. 2001, 2, 835–841. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Palsson-McDermott, E.M.; Bowie, A.G.; Jefferies, C.A.; Mansell, A.S.; Brady, G.; Brint, E.; Dunne, A.; Gray, P.; Harte, M.T.; et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nat. Cell Biol. 2001, 413, 78–83. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Im, E.; Chung, H.K.; Pothoulakis, C.; Rhee, S.H. TRIF mediates toll-like receptor 5-induced signaling in intestinal epithelial cells. J. Biol. Chem. 2010, 285, 37570–37578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, S.L.; Jefferies, C.A.; Feighery, C.; O’Neill, L.A. Signaling by Toll-like receptors 8 and 9 requires Bruton’s tyrosine kinase. J. Biol. Chem. 2007, 282, 36953–36960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Wang, M.; Qi, J.; Wang, H.; Li, X.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006, 281, 5895–5907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Xiong, Z.; Xu, X.; Li, F.; Lu, L.; Li, W.; Su, J.; Liu, Y.; Liu, D.; Xie, Z.; et al. Loss-of-function mutations in filaggrin gene associate with psoriasis vulgaris in Chinese population. Qual. Life Res. 2012, 131, 1269–1274. [Google Scholar] [CrossRef]
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef] [Green Version]
- Grželj, J.; Mlinarič-Raščan, I.; Marko, P.B.; Marovt, M.; Gmeiner, T.; Šmid, A. Polymorphisms in GNMT and DNMT3b are associated with methotrexate treatment outcome in plaque psoriasis. Biomed. Pharmacother. 2021, 138, 111456. [Google Scholar] [CrossRef]
- Zhou, B.; Wei, F.Y.; Kanai, N.; Fujimura, A.; Kaitsuka, T.; Tomizawa, K. Identification of a splicing variant that regulates type 2 di-abetes risk factor CDKAL1 level by a coding-independent mechanism in human. Hum. Mol. Genet. 2014, 23, 4639–4650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 in-fluences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coto-Segura, P.; Batalla, A.; González-Fernández, D.; Gomez, J.; Santos-Juanes, J.; Queiro, R.; Alonso, B.; Iglesias, S.; Coto, E. CDKAL1 gene variants affect the anti-TNF response among Psoriasis patients. Int. Immunopharmacol. 2015, 29, 947–949. [Google Scholar] [CrossRef]
- Bertin, J.; Wang, L.; Guo, Y.; Jacobson, M.D.; Poyet, J.-L.; Srinivasula, S.M.; Merriam, S.; DiStefano, P.S.; Alnemri, E.S. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-κB. J. Biol. Chem. 2001, 276, 11877–11882. [Google Scholar] [CrossRef]
- Coto-Segura, P.; González-Fernández, D.; Batalla, A.; Gómez, J.; González-Lara, L.; Queiro, R.; Alonso, B.; Iglesias, S.; Coto, E. Common and rare CARD14 gene variants affect the antitumour necrosis factor response among patients with psoriasis. Br. J. Dermatol. 2016, 175, 134–141. [Google Scholar] [CrossRef] [PubMed]


| Gene | SNP | Year | N | Population | Drugs | Response | Results | Responsive Allele or Genotype | PMID | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (Months) | Outcome Measure | OR | CI95% | p-value | ||||||||
| Haplotype HLA-A/ TRAF3IP2 | rs9260313/ rs13190932 | 2018 | 109 | Spain | INF ADA | 6 | PASI75 | - | - | <0.05 a | GT | 28921458 |
| Haplotype HLA-B | HLA-B*46 | 2012 | 74 | China | ETN | 3 | PASI50 | - | - | 1 b | - | 21985130 |
| UTK | 0.32 b | |||||||||||
| HLA-B/MICA | rs13437088 | 2017 | 81 | Spain | ETN | 3 | PASI75 | 589.99 | 2.71–128,614.40 | 0.02 | TT | 28470127 |
| HLA-C | rs12191877 | 2016 | 144 | Spain | Anti-TNF | 3 | PASI75 | 0.30 | 0.11–0.88 | 0.027 | T | 27670765 |
| rs1048554 | 2016 | 250 | Greece | Anti-TNF | 6 | PASI75 | 3.94 | 1.16–13.3 | 0.0032 | C | 27043841 | |
| rs610604 | ADA | 6 | PASI75 | - | - | 0.05 b | A | |||||
| - | 2013 | 109 | Spain | Anti-TNF | 6 | PASI75 | 85.1 | 71.7–93.8 | 0.049 | (-) | 23662788 | |
| - | 2013 | 123 | Italy | ETN INF | 3 | PASI75 | - | - | >0.05 | (+) | - | |
| - | 2014 | 138 | United Kingdom and Ireland | ADA ETN | 6 | PASI75 | - | - | >0.05 | (+) | 24758522 | |
| HLA-Cw*06/ LCE3C_ LCE3B del/ins | 2015 | 116 | Spain | Anti-TNF | 3 | PASI75 | 3.14 | 1.07–9.24 | 0.034 | (+)/del | 25794162 | |
| - | 2016 | 96 | Italy | ETN | 3 | PASI75 | - | - | >0.05 | (+) | 27348478 | |
| - | 2017 | 122 | Italy | ADA | 3 | PASI75 | 1.11 | 0.52–2.36 | 0.78 | (+) | 28130758 | |
| - | 2013 | 51 | Italy | UTK | 1 | PASI75 | 5.36 | 1.24–23.1 | 0.024 | (+) | 23521149 | |
| 3 | PASI75 | 13.4 | 1.6–12.6 | <0.008 | ||||||||
| 3 | PASI90 | 4.6 | - | 0.02 | ||||||||
| 10 | PASI75 | 3.9 | 2–7.37 | 0.014 | ||||||||
| 10 | PASI90 | 8.7 | - | 0.012 | ||||||||
| - | 2016 | 134 | Italy | UTK | 3 | PASI75 | 4.1 | - | 0.001 | (+) | 26775778 | |
| 13 | PASI75 | 3.7 | - | 0.003 | ||||||||
| - | 2017 | 255 | Belgium, Italy, Netherlands | UTK | 3 | PASI75 | 3.28 | 1.92–5.59 | <0.001 | (+) | 28207934 | |
| 13 | PASI75 | 3.82 | 1.88–7.73 | <0.001 | ||||||||
| - | 2014 | 66 | China | UTK | 4 | PASI75 | 0.28 | 0.11–0.68 | 0.005 | (+) | 24734995 | |
| - | 2016 | 332 | United States | UTK | 3 | PASI75 | - | - | <0.05 b | (+) | 27476722 | |
| HLA-C*06:02 (HLA-Cw*06 | HLA-Cw6/L12B rs3212227 | 2016 | 64 | Italy | UTK | 1 | PASI75 | 10.49 | 50–0 | 0.009 | (+)/C | 26678060 |
| 13 | 5.21 | 83.3–44.4 | 0.007 | |||||||||
| HLA-Cw6/IL12B rs6887695 | 1 | PASI75 | 6.11 | 23.5–10.5 | 0.031 | (+)/GG | ||||||
| 13 | 4.75 | 82.4–42.1 | 0.006 | |||||||||
| HLA-Cw6 y IL6 rs1800795 | 1 | PASI75 | 6.52 | 38.5–0 | 0.027 | (+)/C | ||||||
| 13 | 4.99 | 84.9–46.7 | 0.005 | |||||||||
| - | 2019 | 1048 | Caucasians and Asian | UTK | 6 | PASI75 | 0.24 | 0.14–0.35 | <0.001 | (+) | 30994858 | |
| - | 2019 | 1326 | United Kingdom and Ireland | ADA | 6 | PASI90 | 2.95 | - | <0.001 | (-) | 30578879 | |
| HLA-G 14-pb ins/del | rs66554220 | 2014 | 11 | Italy | Anti-TNF | 4 | PASI75 | - | - | 0.7 b | (+) | 24909182 |
| Gene | SNP | Year | N | Population | Pathology | Drugs | Response | Results | Responsive Allele or Genotype | PMID | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (Months) | Outcome Measure | OR | CI95% | p-value | |||||||||
| TNF-α-238 | rs361525 | 2013 | 109 | Spain | PS | Anti-TNF | 6 | PASI75 | - | - | 0.049 f | G | 23662788 |
| TNF-α-857 | rs1799724 | - | BSA | - | - | 0.004 f | C | ||||||
| - | PASI | - | - | 0.009 f | |||||||||
| 6 | PASI75 | - | - | 0.006 f | |||||||||
| TNF-α-1031 | rs1799964 | 3 | PASI75 | - | - | 0.047 f | TT | ||||||
| 6 | 0.038 f | ||||||||||||
| TNF-α-308 | rs1800629 | 2015 | 807 | Caucasians | AE IBD APS PS | Anti-TNF | - | - | 2.005 | 1.417–2.838 | 0.000086 | G | 26244882 |
| TNF-α-238 | rs361525 | 500 | 2.196 | 1161–4.154 | 0.016 | G | |||||||
| TNF-α-857 | rs1799724 | 483 | 1.779 | 1.13–2.802 | 0.013 | C | |||||||
| 177 | PS | ETN | - | - | 2.238 | 1.319–3.798 | 0.003 | C | |||||
| IL1-β | rs1143623 | 2017 | 376 | Denmark | PS | Anti-TNF | 3 | PASI75 | 0.35 | - | 0.0041 | GG | 28696418 |
| 230 | UTK | 0.25 | - | 0.0049 | |||||||||
| rs1143627 | 2017 | 376 | PS | Anti-TNF | 3 | PASI75 | 0.28 | - | 0.0016 | AA | |||
| 230 | UTK | 0.24 | - | 0.0042 | |||||||||
| IL6 | rs1800795 | 2012 | 60 | Italy | PS | Anti-TNF | 6 | PASI75 | 2.00 | 1.19–3.38 | ≤0.05 | GG | 22158445 |
| IL12β | rs2546890 | 2017 | 78 | Spain | PS APS | ETN | 6 | PASI75 | 11.92 | 1.07–132.67 | 0.044 | G | 28470127 |
| 2017 | 144 | Spain | PS | Anti-TNF | 3 | PASI75 | 3.22 | 1.23–8.40 | 0.017 | G | 27670765 | ||
| 6 | 4.14 | 1.23–13.81 | 0.022 | ||||||||||
| 12 | 2.79 | 1.02–7.64 | 0.046 | ||||||||||
| rs3213094 | 2017 | 66 | Netherlands | PS | UTK | 3 | ΔPASI | −3.15 c | −5.724– −0.586 | 0.017 | CT | 27564082 | |
| IL17F | rs763780 | 2015 | 67 | Spain | PS | UTK | 4 | PASI75 | 12.23 | 1.17–127.36 | 0.022 | CT | 26347322 |
| 7 | 14.18 | 1.35–149.42 | 0.016 | ||||||||||
| 62 | ADA | 7 | PASI75 | 14.00 | 2.15–91.12 | 0.0044 | |||||||
| TNFAIP3 | rs610604 | 2013 | 51 | Italy | PS | UTK | 10 | PASI75 | 1.6 | - | 0.75 | - | 23521149 |
| 2016 | 64 | Italy | PS | UTK | 3 | PASI75 | - | - | >0.05 | - | 26678060 | ||
| 2017 | 66 | Netherlands | PS | UTK | 3 | ΔPASI | 3.490 c | 0.329–6.650 | 0.031 | GG | 27564082 | ||
| PS APS | 11.230 c | 7.486–14.973 | <0.001 | ||||||||||
| 2019 | 20 | Spain | PS APS | Anti-TNF | 3 | % EQ-VAS | −10.60 | −20.71–0.048 | 0.041 | AC/CC | 30653751 | ||
| rs6920220 | −25.83 | −47.969– −3.698 | 0.025 | AA | |||||||||
| PDE3A-SLCO1C1 | rs11045392-rs3794271 | 2015 | 130 | Spain | PS | Anti-TNF | 3 | ΔPASI | - | - | 0.00057 | G | 25403996 |
| TNFRSF1B | rs1061622 | 2012 | 80 | Greece | PS | Anti-TNF | 6 | PASI75 | - | - | 0.019 f | TT | 22111980 |
| ETN | - | - | 0.001 f | ||||||||||
| 2015 | 90 | Spain | PS | Anti-TNF | 6 | PASI50 | 2.96 | 1.09–8.02 | 0.03 | G | 25537528 | ||
| 2015 | 929 | Caucasians and Asian | RA CD PS | Anti-TNF | - | - | 0.72 | 0.57–0.93 | 0.01 | T | 26071216 | ||
| 2015 | 170 | PS | - | - | 0.39 | 0.23–0.67 | <0.001 | T | |||||
| CD84 | rs6427528 | 2013 | 733 | Caucasians | RA | ETN | - | - | - | - | 0.004 | AG | 23555300 |
| 2017 | 161 | Netherlands | PS | ETN | 3 | ΔPASI | −2.028 c | −3.794–0.261 | 0.025 | AG | 27564082 | ||
| FCGR2A | rs1801274 | 2013 | 70 | Spain | PS | Anti-TNF | 6a | dBSA | - | - | 0.03 d | 131HH | 24048425 |
| 2015 | 115 | Spain | PS | Anti-TNF | 6 | PASI75 | - | - | 0.1 e | - | 26398016 | ||
| 2016 | 100 | Greece | PS | Anti-TNF | 6 | PASI75 | - | - | 0.882 e | H131R | 27044681 | ||
| 2016 | 133 | Spain | PS | Anti-TNF | 6a | PASI75 | 13.32 | 1.67–106.50 | 0.015 | 131RR | 27670765 | ||
| FCGR3A | rs396991 | 2005 | 35 | United States | PS RA | Anti-TNF | 6a | - | - | - | <0.01 f | 158FF | 16142749 |
| 2013 | 70 | Spain | PS | Anti-TNF | 6a | dBSA | - | - | 0.02 d | 158FF | 24048425 | ||
| 2015 | 115 | Spain | PS | Anti-TNF | 6 | PASI75 | 12.05 | 1.25–111.11 | 0.04 | 158FF | 26398016 | ||
| 2016 | 100 | Greece | PS | Anti-TNF | 6 | PASI75 | 2.96 | 1.601–5.527 | 0.0018 | 158 V | 27044681 | ||
| IL17RA | rs4819554 | 2018 | 238 | Spain | PS | Anti-TNF | 3 | PASI75 | 1.86 | 1.05–3.27 | 0.03 | A | 27670766 |
| IL23R | rs11209026 | 2013 | 109 | Spain | PS | Anti-TNF | 6 | PASI90 | - | - | 0.006f | GG | 23662788 |
| TLR2 | rs4696480 | 2017 | 376 | Denmark | PS | Anti-TNF | 3 | PASI75 | 0.22 | 0.08–0.59 | 0.0032 | A | 28696418 |
| rs11938228 | 0.30 | 0.14–0.64 | 0.0019 | C | |||||||||
| LY96 | rs11465996 | 230 | UTK | 3 | ΔPASI | 0.33 | 0.15–0.71 | 0.0044 | C | ||||
| TIRAP | rs8177374 | 230 | 3 | PASI75 | 9.42 | 1.96–45.3 | 0.0051 | C | |||||
| TLR5 | rs5744174 | 3 | ΔPASI | 5.26 | 1.93–14.38 | 0.0012 | A | ||||||
| TLR9 | rs352139 | 376 | Anti-TNF | 225b | DS | 2.42 | 1.32–4.44 | 0.0044 | G | ||||
| PGLYR4-24 | rs2916205 | 2016 | 144 | Spain | PS | Anti-TNF | 3 | PASI75 | 3.62 | 1.00–13.07 | 0.05 | C | 27670765 |
| CDKAL1 | rs6908425 | 2015 | 116 | Spain | PS | Anti-TNF | 6 | PASI75 | 3.14 | 1.40–7.05 | 0.005 | CC | 26563541 |
| 2016 | 133 | Spain | PS | Anti-TNF | 6 | PASI75 | 0.14 | 0.03–0.66 | 0.013 | T | 27670765 | ||
| CARD14 | rs11652075 | 2016 | 116 | Spain | PS | Anti-TNF | 6 | PASI75 | 3.71 | 1.30–10.51 | 0.01 | CC | 26854129 |
| PTTG1 | rs2431697 | 2017 | 78 | Spain | PS APS | ETN | 3 | PASI75 | 29.80 | 1.16–765.68 | 0.04 | C | 28470127 |
| MAP3K1 | rs96844 | 2017 | 78 | Spain | PS APS | ETN | 3 | PASI75 | 0.01 | 0–0.33 | 0.009 | C | 28470127 |
| 2016 | 144 | Spain | PS | Anti-TNF | 3 | PASI75 | 0.17 | 0.05–0.56 | 0.017 | C | 27670765 | ||
| 6 | PASI75 | 0.24 | 0.06–0.97 | 0.045 | |||||||||
| ZNF816A | rs9304742 | 2017 | 78 | Spain | PS APS | ETN | 3 | PASI75 | 8144.11 | 13.03–5089337.0 | 0.006 | CC | 28470127 |
| 2016 | 144 | Spain | PS | Anti-TNF | 3 | PASI75 | 7.66 | 1.37–42.70 | 0.02 | CC | 27670765 | ||
| GBP6 | rs928655 | 2017 | 68 | Spain | PS APS | ETN | 6 | PASI75 | 0.14 | 0.03–0.67 | 0.014 | G | 28470127 |
| CTNNA2 | rs11126740 | 2016 | 144 | Spain | PS | Anti-TNF | 3 | PASI75 | 20.56 | 2.75–153.69 | 0.003 | AA | 27670765 |
| HTR2A | rs6311 | 6 | PASI75 | 5.6 | 1.10–28.63 | 0.038 | T | ||||||
| Gen | SNP | Year | N | Results | Responsive Allele or Genotype | PMID | ||
|---|---|---|---|---|---|---|---|---|
| OR | CI95% | p-value | ||||||
| CTLA4 | rs3087243 | 2016 | 161 | 0.001 | 0–0.24 | 0.012 | AG/GG | 26194362 |
| FBXL19 | rs10782001 | 32.85 | 1.46–738.37 | 0.0028 | GG | |||
| IL23R | rs11209026 | 11,011.59 | 17.36–6984187.8 | 0.005 | AG | |||
| SLC12A8 | rs651630 | 0 | 0–0.06 | 0.011 | AA | |||
| TAP1 | rs1800453 | 0.009 | 0–0.45 | 0.018 | AG | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Membrive Jiménez, C.; Pérez Ramírez, C.; Sánchez Martín, A.; Vieira Maroun, S.; Arias Santiago, S.A.; Ramírez Tortosa, M.d.C.; Jiménez Morales, A. Influence of Genetic Polymorphisms on Response to Biologics in Moderate-to-Severe Psoriasis. J. Pers. Med. 2021, 11, 293. https://doi.org/10.3390/jpm11040293
Membrive Jiménez C, Pérez Ramírez C, Sánchez Martín A, Vieira Maroun S, Arias Santiago SA, Ramírez Tortosa MdC, Jiménez Morales A. Influence of Genetic Polymorphisms on Response to Biologics in Moderate-to-Severe Psoriasis. Journal of Personalized Medicine. 2021; 11(4):293. https://doi.org/10.3390/jpm11040293
Chicago/Turabian StyleMembrive Jiménez, Cristina, Cristina Pérez Ramírez, Almudena Sánchez Martín, Sayleth Vieira Maroun, Salvador Antonio Arias Santiago, María del Carmen Ramírez Tortosa, and Alberto Jiménez Morales. 2021. "Influence of Genetic Polymorphisms on Response to Biologics in Moderate-to-Severe Psoriasis" Journal of Personalized Medicine 11, no. 4: 293. https://doi.org/10.3390/jpm11040293
APA StyleMembrive Jiménez, C., Pérez Ramírez, C., Sánchez Martín, A., Vieira Maroun, S., Arias Santiago, S. A., Ramírez Tortosa, M. d. C., & Jiménez Morales, A. (2021). Influence of Genetic Polymorphisms on Response to Biologics in Moderate-to-Severe Psoriasis. Journal of Personalized Medicine, 11(4), 293. https://doi.org/10.3390/jpm11040293