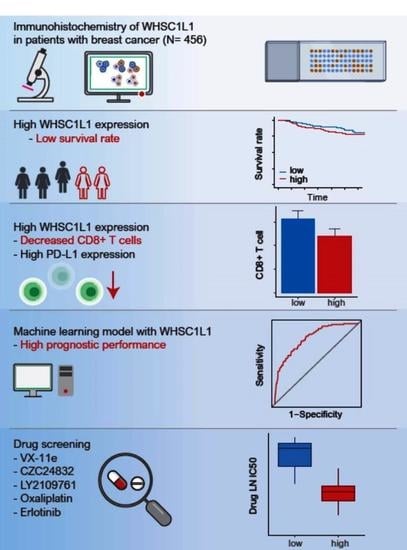
High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Tissue Microarray Construction and Immunohistochemistry in Our Cohort
2.3. Analysis Based on the METABRIC Database and TCGA Database
2.4. Machine Learning Algorithm for Validation
2.5. GDSC Database
2.6. Statistical Analysis
3. Results
3.1. Clinicopathological Parameters and Survival Rate
3.2. Anticancer Immune Response and Pathway Network Analysis
3.3. Machine Learning and Drug Screening
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.B.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, B.; Yang, J.; Wang, H.; Yang, G.; Xu, R.; You, L.; Zhao, Y. The role of histone methylation in the development of digestive cancers: A potential direction for cancer management. Signal. Transduct. Target Ther. 2020, 5, 143. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.J.; Carpenter, P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Bio. 2012, 13, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.X.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget 2015, 6, 2466–2482. [Google Scholar] [CrossRef] [Green Version]
- Angrand, P.O.; Apiou, F.; Stewart, A.F.; Dutrillaux, B.; Losson, R.; Chambon, P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 2001, 74, 79–88. [Google Scholar] [CrossRef]
- Kang, D.; Cho, H.S.; Toyokawa, G.; Kogure, M.; Yamane, Y.; Iwai, Y.; Hayami, S.; Tsunoda, T.; Field, H.I.; Matsuda, K.; et al. The histone methyltransferase Wolf-Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Gene Chromosome Canc. 2013, 52, 126–139. [Google Scholar] [CrossRef]
- Mahmood, S.F.; Gruel, N.; Nicolle, R.; Chapeaublanc, E.; Delattre, O.; Radvanyi, F.; Bernard-Pierrot, I. PPAPDC1B and WHSC1L1 Are Common Drivers of the 8p11-12 Amplicon, Not Only in Breast Tumors But Also in Pancreatic Adenocarcinomas and Lung Tumors. Am. J. Pathol. 2013, 183, 1634–1644. [Google Scholar] [CrossRef]
- Yang, Z.Q.; Liu, G.; Bollig-Fischer, A.; Giroux, C.N.; Ethier, S.P. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010, 70, 8487–8497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Thomsen, R.; Kahns, S.; Nielsen, A.L. The NSD3L histone methyltransferase regulates cell cycle and cell invasion in breast cancer cells. Biochem. Biophys. Res. Commun. 2010, 398, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.C.; Mills, J.N.; Turner-Ivey, B.; Wilson, R.C.; Guest, S.T.; Rutkovsky, A.; Dombkowski, A.; Kappler, C.S.; Hardiman, G.; Ethier, S.P. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ER alpha in SUM-44 breast cancer cells and is associated with ER alpha over-expression in breast cancer. Mol. Oncol. 2016, 10, 850–865. [Google Scholar] [CrossRef]
- Jeong, G.Y.; Park, M.K.; Choi, H.J.; An, H.W.; Park, Y.U.; Choi, H.J.; Park, J.; Kim, H.Y.; Son, T.; Lee, H.; et al. NSD3-Induced Methylation of H3K36 Activates NOTCH Signaling to Drive Breast Tumor Initiation and Metastatic Progression. Cancer Res. 2021, 81, 77–90. [Google Scholar] [CrossRef]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum. Cell 2020, 33, 930–937. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Brennan, K.; Shin, J.H.; Tay, J.K.; Prunello, M.; Gentles, A.J.; Sunwoo, J.B.; Gevaert, O. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci. Rep. 2017, 7, 17064. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, G.W.; Wang, K.; Xia, Y.X.; Wang, J.S.; Wang, X.H.; Li, X.C. Integrating Machine Learning and Tumor Immune Signature to Predict Oncologic Outcomes in Resected Biliary Tract Cancer. Ann. Surg. Oncol. 2020. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [Green Version]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Sauerbrei, W.; Taube, S.E.; McShane, L.M.; Cavenagh, M.M.; Altman, D.G. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. J. Natl. Cancer Inst. 2018, 110, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Remmele, W.; Stegner, H.E. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue]. Pathologe 1987, 8, 138–140. [Google Scholar]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [Green Version]
- El-Nachef, D.; Oyama, K.; Wu, Y.Y.; Freeman, M.; Zhang, Y.; MacLellan, W.R. Repressive histone methylation regulates cardiac myocyte cell cycle exit. J. Mol. Cell. Cardiol. 2018, 121, 1–12. [Google Scholar] [CrossRef]
- Haydn, T.; Metzger, E.; Schuele, R.; Fulda, S. Concomitant epigenetic targeting of LSD1 and HDAC synergistically induces mitochondrial apoptosis in rhabdomyosarcoma cells. Cell Death Dis. 2017, 8, e2879. [Google Scholar] [CrossRef]
- Higgs, M.R.; Sato, K.; Reynolds, J.J.; Begum, S.; Bayley, R.; Goula, A.; Vernet, A.; Paquin, K.L.; Skalnik, D.G.; Kobayashi, W.; et al. Histone Methylation by SETD1A Protects Nascent DNA through the Nucleosome Chaperone Activity of FANCD2. Mol. Cell. 2018, 71, 25–41.e26. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Nayak, S.; Jankowitz, R.; Davidson, N.E.; Oesterreich, S. Epigenetics in breast cancer: what’s new? Breast Cancer Res. 2011, 13, 225. [Google Scholar] [CrossRef] [Green Version]
- Gelsi-Boyer, V.; Orsetti, B.; Cervera, N.; Finetti, P.; Sircoulomb, F.; Rouge, C.; Lasorsa, L.; Letessier, A.; Ginestier, C.; Monville, F.; et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol. Cancer Res. 2005, 3, 655–667. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Li, F.D.; Zhang, J.H.; Wu, J.H.; Shi, Y.Y. The Methyltransferase NSD3 Has Chromatin-binding Motifs, PHD5-C5HCH, That Are Distinct from Other NSD (Nuclear Receptor SET Domain) Family Members in Their Histone H3 Recognition. J. Biol. Chem. 2013, 288, 4692–4703. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Trojer, P.; Xu, C.F.; Cheung, P.; Kuo, A.; Drury, W.J., 3rd; Qiao, Q.; Neubert, T.A.; Xu, R.M.; Gozani, O.; et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 2009, 284, 34283–34295. [Google Scholar] [CrossRef] [Green Version]
- Morishita, M.; Mevius, D.; di Luccio, E. In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct. Biol. 2014, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Turner-Ivey, B.; Smith, E.L.; Rutkovsky, A.C.; Spruill, L.S.; Mills, J.N.; Ethier, S.P. Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3. Breast Cancer Res. Treat. 2017, 164, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Piao, L.; Zhuang, M.; Qiu, X.; Xu, X.; Zhang, D.; Liu, M.; Ren, D. Silencing of histone methyltransferase NSD3 reduces cell viability in osteosarcoma with induction of apoptosis. Oncol. Rep. 2017, 38, 2796–2802. [Google Scholar] [CrossRef] [Green Version]
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Deng, X.; Kiyotani, K.; Park, J.H.; Matsuo, Y.; Lingen, M.; Suzuki, T.; Dohmae, N.; et al. WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci. Rep. 2017, 7, 40664. [Google Scholar] [CrossRef]
- Yi, L.; Yi, L.; Liu, Q.; Li, C. Downregulation of NSD3 (WHSC1L1) inhibits cell proliferation and migration via ERK1/2 deactivation and decreasing CAPG expression in colorectal cancer cells. Onco. Targets Ther. 2019, 12, 3933–3943. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Jasek-Gajda, E.; Jurkowska, H.; Jasinska, M.; Litwin, J.A.; Lis, G.J. Combination of ERK2 inhibitor VX-11e and voreloxin synergistically enhances anti-proliferative and pro-apoptotic effects in leukemia cells. Apoptosis 2019, 24, 849–861. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Guo, X.; Zhang, H.; Kong, X.; Yang, F.; Zheng, C. Mechanism of action and efficacy of LY2109761, a TGF-beta receptor inhibitor, targeting tumor microenvironment in liver cancer after TACE. Oncotarget 2018, 9, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Ladygina, N.; Gottipati, S.; Ngo, K.; Castro, G.; Ma, J.Y.; Banie, H.; Rao, T.S.; Fung-Leung, W.P. PI3K gamma kinase activity is required for optimal T-cell activation and differentiation. Eur. J. Immunol. 2013, 43, 3183–3196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.; Qu, X.; Fan, Y.; Che, X.; Qu, J.; Xu, L.; Liu, J.; Liu, Y. Trastuzumab and oxaliplatin exhibit a synergistic antitumor effect in HER2-postive gastric cancer cells. Anticancer. Drugs 2014, 25, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Gridelli, C.; Rossi, A.; Ciardiello, F.; De Marinis, F.; Crino, L.; Morabito, A.; Morgillo, F.; Montanino, A.; Daniele, G.; Piccirillo, M.C.; et al. BEVERLY: Rationale and Design of a Randomized Open-Label Phase III Trial Comparing Bevacizumab Plus Erlotinib Versus Erlotinib Alone as First-Line Treatment of Patients With EGFR-Mutated Advanced Nonsquamous Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2016, 17, 461–465. [Google Scholar] [CrossRef] [PubMed]





| Parameter | WHSC1L1 (HYGH Cohort) | p-Value | |
|---|---|---|---|
| Low (n = 224), n (%) | High (n = 232), n (%) | ||
| Age | 49.5 ± 9.3 | 49.5 ± 10.5 | 0.99 1 |
| T stage | |||
| 1 | 109 (48.7) | 99 (42.7) | 0.06 3 |
| 2 | 109 (48.7) | 117 (50.4) | |
| 3 | 6 (2.7) | 16 (6.9) | |
| N stage | |||
| 0 | 115 (51.3) | 118 (50.9) | 0.763 3 |
| 1 | 64 (28.6) | 71 (30.6) | |
| 2 | 31 (13.8) | 29 (12.5) | |
| 3 | 14 (6.2) | 14 (6.0) | |
| Histological grade | |||
| 1 | 39 (17.4) | 48 (20.7) | 0.335 3 |
| 2 | 115 (51.3) | 101 (43.5) | |
| 3 | 70 (31.2) | 83 (35.8) | |
| Lymphatic invasion | |||
| Negative | 123 (54.9) | 107 (46.1) | 0.075 1 |
| Positive | 101 (45.1) | 125 (53.9) | |
| Vascular invasion | |||
| Negative | 213 (95.1) | 213 (91.8) | 0.221 1 |
| Positive | 11 (4.9) | 19 (8.2) | |
| Perineural invasion | |||
| Negative | 184 (82.1) | 178 (76.7) | 0.189 1 |
| Positive | 40 (17.9) | 54 (23.3) | |
| ER | |||
| Negative | 53 (23.7) | 76 (32.8) | 0.041 |
| Positive | 171 (76.3) | 156 (67.2) | |
| PR | |||
| Negative | 75 (33.5) | 104 (44.8) | 0.0171 |
| Positive | 149 (66.5) | 128 (55.2) | |
| HER2 | |||
| Negative | 161 (71.9) | 157 (67.7) | 0.382 1 |
| Positive | 63 (28.1) | 75 (32.3) | |
| PD-L1 | |||
| Negative | 155 (62.9) | 127 (54.7) | 0.0011 |
| Positive | 69 (30.8) | 105 (45.3) | |
| P53 percentage | 8.4 ± 11.9 | 11.8 ± 13.7 | 0.0052 |
| Ki-67 index | 21.5 ± 32.8 | 34.0 ± 38.5 | <0.0012 |
| Disease-Free Survival | Univariate 1 | Multivariate 2 | HR | 95% CI | |
| WHSC1L1 (low vs. high) | <0.001 | <0.001 | 2.265 | 1.451 | 3.537 |
| T stage (1, 2 vs. 3) | 0.001 | 0.001 | 2.820 | 1.507 | 5.275 |
| N stage (0, 1 vs. 2, 3) | <0.001 | 0.001 | 2.124 | 1.334 | 3.381 |
| Histological grade (1, 2 vs. 3) | 0.01 | 0.05 | 1.529 | 0.988 | 2.368 |
| Lymphatic invasion (absence vs. presence) | <0.001 | 0.234 | 1.353 | 0.822 | 2.225 |
| Perineural invasion (absence vs. presence) | <0.001 | <0.001 | 2.192 | 1.415 | 3.394 |
| Estrogen receptor (negative vs. positive) | 0.045 | 0.05 | 0.647 | 0.414 | 1.011 |
| Disease-specific survival | Univariate 1 | Multivariate 2 | HR | 95% CI | |
| WHSC1L1 (low vs. high) | <0.001 | <0.001 | 2.505 | 1.567 | 4.005 |
| T stage (1, 2 vs. 3) | <0.001 | 0.022 | 2.251 | 1.122 | 4.516 |
| N stage (0, 1 vs. 2, 3) | <0.001 | <0.001 | 2.494 | 1.541 | 4.037 |
| Histological grade (1, 2 vs. 3) | 0.001 | 0.3 | 1.274 | 0.806 | 2.015 |
| Lymphatic invasion (absence vs. presence) | <0.001 | 0.31 | 1.311 | 0.777 | 2.210 |
| Perineural invasion (absence vs. presence) | <0.001 | <0.001 | 2.477 | 1.586 | 3.868 |
| Estrogen receptor (negative vs. positive) | 0.008 | 0.076 | 0.655 | 0.410 | 1.046 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-S.; Min, K.-W.; Kim, D.-H.; Son, B.-K.; Kwon, M.-J.; Hong, S.-M. High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach. J. Pers. Med. 2021, 11, 636. https://doi.org/10.3390/jpm11070636
Kim H-S, Min K-W, Kim D-H, Son B-K, Kwon M-J, Hong S-M. High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach. Journal of Personalized Medicine. 2021; 11(7):636. https://doi.org/10.3390/jpm11070636
Chicago/Turabian StyleKim, Hyung-Suk, Kyueng-Whan Min, Dong-Hoon Kim, Byoung-Kwan Son, Mi-Jung Kwon, and Sang-Mo Hong. 2021. "High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach" Journal of Personalized Medicine 11, no. 7: 636. https://doi.org/10.3390/jpm11070636
APA StyleKim, H. -S., Min, K. -W., Kim, D. -H., Son, B. -K., Kwon, M. -J., & Hong, S. -M. (2021). High WHSC1L1 Expression Reduces Survival Rates in Operated Breast Cancer Patients with Decreased CD8+ T Cells: Machine Learning Approach. Journal of Personalized Medicine, 11(7), 636. https://doi.org/10.3390/jpm11070636