MRI Deep Learning-Based Solution for Alzheimer’s Disease Prediction
Abstract
:1. Introduction
2. Related Work
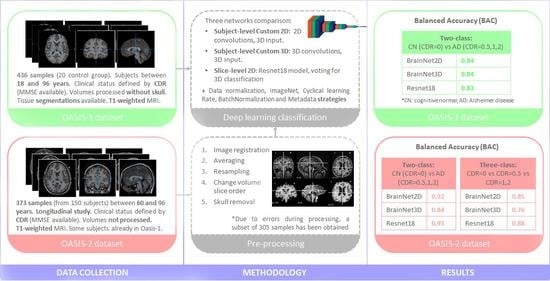
2.1. Data Collections
2.2. Automatic Analysis
3. Materials and Methods
3.1. Dataset
3.2. Data and MRI Volume Processing: Model Input
3.2.1. Data Normalization
3.2.2. Metadata
3.3. Proposed Solution
- BrainNet3D: A custom 3D network is proposed similar to the previous one but with a real 3D approach. Input data are M, N, K and 1, with M and N being the size of every slice, and K the number of slices. Five convolutional blocks are proposed, each of them containing the 3D convolutional layer and 3D max pooling layers. This approach fits better with biology and human understanding and might provide more useful patterns of structures in the brain. Filters for each convolutional layer represent the number of filters ∗filter size, with the number of filters being 8, 16, 32, 64 and 128 in each of the five convolutional blocks, and the filter size is 3 in all of them. At the end of the embedding part, a Global Average Pooling 3D layer is added. No additional fully connected layers are added. The activation function is ‘softmax’ for exclusive classes. The loss function is initially ‘categorical_crossentropy’. The architecture of BrainNet3D can be seen in Figure 1. Again, two variations of the same architecture are compared. First, the baseline BrainNet3D architecture (Figure 2A), and second, including the Batch Normalization layers as an improvement technique (Figure 2B).
- 2D slice-level network: This network architecture performs at the 2D slice-level. This means that the network has as input a single slice of M, N and 3 size, and every image has an associated classification output. The unique channel per slice has been replicated to provide the three-channel input expected by ResNet family networks and to make it possible to fine-tune operations with ImageNet weights. ResNet18, a small model from the ResNet family [45] pretrained on ImageNet, has been used as a reference model. After the embedding part, and as in the previous networks, the activation function is ‘softmax’ and the loss function is ‘categorical_crossentropy’. In the testing phase, it must be pointed out that the final output for a complete study is provided in terms of ‘majority vote’ over all the slices of the sequence.
- Cyclical Learning Rate: Cyclical Learning Rate (CLR) [49] is a strategy that allows oscillating between two learning rate values, iteratively. Previous traditional well-known learning strategies usually consider gradually decreasing the learning rate over the epoch using different functions (linear, polynomial, step, etc.). However, this strategy can lead the model to descend to areas of low loss values. With CLR, the optimal learning rate parameter can be easily found, and the model can consequently be better (and sooner) tuned. A minimum and a maximum learning rate value must be defined, and then the rate will cyclically oscillate between the two bounds. To do so, the different working policies can be defined: “triangular”, which is a simple triangular cycle, “triangular2”, also triangular but additionally cutting the maximum learning rate in half every cycle, and “exp_range”, which is similar to the previous but with an exponential decay. Our experiments with CLR revealed that the triangular policy reported the best results with our datasets.
- Batch Normalization: Batch Normalization [50] is a technique for training deep neural networks that standardizes the inputs to a layer for each mini-batch entering the network. This has the effect of stabilizing the learning process and often contributes to a better training process and thus better performance of the obtained model. Batch Normalization can also help to reduce overfitting, which is one of the main issues whenever a dataset is not large enough, as is the case with OASIS-1 and OASIS-2. This overfitting problem is mainly remarkable in the 3D-subject level approach. For the application of this Batch Normalization, the keras BatchNormalization() function, together with a ‘relu’ activation, is applied after the convolutional layer and before the max pooling layer. Figure 1 and Figure 2 illustrate how these layers have been integrated in the proposed BrainNet2D and BrainNet3D architectures, respectively.
- Metadata: Sex and age have been revealed as relevant variables for the diagnosis, as concluded in Appendix A. Therefore, these two variables could facilitate the establishment of the classification output and have been incorporated in the network [48]. The inclusion in the network has been carried out after the embeddings and before the final activation layer. The metadata has been considered as follows: two classes for sex (male, female), and two classes for age (<60 and >60).
- ImageNet: Some experiments, particularly the ones adopting the ResNet18 architecture, have used pre-trained ImageNet weights. This practice often improves the results and accelerates the training process.
4. Results
5. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Statistical Analysis of Demographics and Clinical Information

Appendix B. Pre-Processing of Raw MRI Volumes
- Image registration, using the Flirt tool from the AFNI library: all images included in the RAW folder of the OASIS-2 database are registered in pairs.
- Averaging registered images, using the fslmaths tool from the FSL library: all resultant images from the registration process are averaged to obtain one unique image.
- Image resampling, using the 3dRESAMPLE tool from the AFNI library: the resultant averaged image is resampled to obtain 1 × 1 × 1 mm voxels.
- Change volume slice order, using the fslswapdim tool from the FSL library: As original images do not have the same slice anatomical plane view as the pre-processed volumes in OASIS-1 and the aim of the pre-processing part is to replicate as well as possible the pre-processing performed in OASIS-1, the dimension slices of the resampled image are swapped.

References
- Smith, M.A. Alzheimer disease. Int. Rev. Neurobiol. 1998, 42, 1–54. [Google Scholar] [CrossRef]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. ‘Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Mini-Mental State Examination Second Edition|MMSE-2. Available online: https://www.parinc.com/Products/Pkey/238 (accessed on 24 March 2021).
- Hughes, C.P.; Berg, L.; Danziger, W.L.; Coben, L.A.; Martin, R.L. A new clinical scale for the staging of dementia. Br. J. Psychiatry 1982, 140, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Duara, R.; Loewenstein, D.A.; Greig-Custo, M.T.; Raj, A.; Barker, W.; Potter, E.; Schofield, E.; Small, B.; Schinka, J.; Wu, Y.; et al. Diagnosis and staging of mild cognitive impairment, using a modification of the clinical dementia rating scale: The mCDR. Int. J. Geriatr. Psychiatry 2010, 25, 282–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, T.K. Clinical Diagnosis of Alzheimer’s Disease. In Biomarkers in Alzheimer’s Disease; Elsevier: Amsterdam, The Netherlands, 2016; pp. 27–48. [Google Scholar]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McRobbie, D.W.; Moore, E.A.; Graves, M.J.; Prince, M.R. MRI from Picture to Proton, 3rd ed.; Cambridge University Press: Cambridge, UK, 2017; ISBN 9781107643239. [Google Scholar]
- Corriveau-Lecavalier, N.; Mellah, S.; Clément, F.; Belleville, S. Evidence of parietal hyperactivation in individuals with mild cognitive impairment who progressed to dementia: A longitudinal fMRI study. NeuroImage Clin. 2019, 24, 101958. [Google Scholar] [CrossRef]
- Dubois, B.; Villain, N.; Frisoni, G.B.; Rabinovici, G.D.; Sabbagh, M.; Cappa, S.; Bejanin, A.; Bombois, S.; Epelbaum, S.; Teichmann, M.; et al. Clinical diagnosis of Alzheimer’s disease: Recommendations of the International Working Group. Lancet Neurol. 2021, 20, 484–496. [Google Scholar] [CrossRef]
- ADNI|Alzheimer’s Disease Neuroimaging Initiative. Available online: http://adni.loni.usc.edu/ (accessed on 23 March 2021).
- Marinescu, R.V.; Oxtoby, N.P.; Young, A.L.; Bron, E.E.; Toga, A.W.; Weiner, M.W.; Barkhof, F.; Fox, N.C.; Klein, S.; Alexander, D.C. TADPOLE challenge: Prediction of longitudinal evolution in Alzheimer’s disease. arXiv 2018, arXiv:1805.03909. [Google Scholar]
- OASIS Brains—Open Access Series of Imaging Studies. Available online: https://www.oasis-brains.org/ (accessed on 23 March 2021).
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef] [Green Version]
- Marcus, D.S.; Fotenos, A.F.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open access series of imaging studies: Longitudinal MRI data in nondemented and demented older adults. J. Cogn. Neurosci. 2010, 22, 2677–2684. [Google Scholar] [CrossRef]
- LaMontagne, P.J.; Benzinger, T.L.S.; Morris, J.C.; Keefe, S.; Hornbeck, R.; Xiong, C.; Grant, E.; Hassenstab, J.; Moulder, K.; Vlassenko, A.G.; et al. OASIS-3: Longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. medRxiv 2019, 12, 13. [Google Scholar] [CrossRef] [Green Version]
- Alzheimer’s Disease Connectome Project. Available online: https://www.humanconnectome.org/study/alzheimers-disease-connectome-project (accessed on 24 March 2021).
- Connectome—Homepage. Available online: https://www.humanconnectome.org/ (accessed on 24 March 2021).
- Bron, E.E.; Smits, M.; van der Flier, W.M.; Vrenken, H.; Barkhof, F.; Scheltens, P.; Papma, J.M.; Steketee, R.M.E.; Méndez Orellana, C.; Meijboom, R.; et al. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: The CADDementia challenge. Neuroimage 2015, 111, 562–579. [Google Scholar] [CrossRef] [Green Version]
- Malone, I.B.; Cash, D.; Ridgway, G.R.; MacManus, D.G.; Ourselin, S.; Fox, N.C.; Schott, J.M. MIRIAD-Public release of a multiple time point Alzheimer’s MR imaging dataset. Neuroimage 2013, 70, 33–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beekly, D.L.; Ramos, E.M.; Lee, W.W.; Deitrich, W.D.; Jacka, M.E.; Wu, J.; Hubbard, J.L.; Koepsell, T.D.; Morris, J.C.; Kukull, W.A.; et al. The National Alzheimer’s Coordinating Center (NACC) database: The uniform data set. Alzheimer Dis. Assoc. Disord. 2007, 21, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, E.; Ballerini, L.; Hernandez, M.D.C.V.; Chappell, F.M.; González-Castro, V.; Anblagan, D.; Danso, S.; Muñoz-Maniega, S.; Job, D.; Pernet, C.; et al. Machine learning of neuroimaging for assisted diagnosis of cognitive impairment and dementia: A systematic review. Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit. 2018, 10, 519–535. [Google Scholar] [CrossRef]
- Vieira, S.; Pinaya, W.H.L.; Mechelli, A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neurosci. Biobehav. Rev. 2017, 74, 58–75. [Google Scholar] [CrossRef] [Green Version]
- Tanveer, M.; Richhariya, B.; Khan, R.U.; Rashid, A.H.; Khanna, P.; Prasad, M.; Lin, C.T. Machine Learning Techniques for the Diagnosis of Alzheimer’s Disease. ACM Trans. Multimed. Comput. Commun. Appl. 2020, 16, 1–35. [Google Scholar] [CrossRef]
- Rathore, S.; Habes, M.; Iftikhar, M.A.; Shacklett, A.; Davatzikos, C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. Neuroimage 2017, 155, 530–548. [Google Scholar] [CrossRef]
- Khagi, B.; Kwon, G.; Lama, R. Comparative analysis of Alzheimer’s disease classification by CDR level using CNN, feature selection, and machine-learning techniques. Int. J. Imaging Syst. Technol. 2019, 29, 297–310. [Google Scholar] [CrossRef]
- Nawaz, H.; Maqsood, M.; Afzal, S.; Aadil, F.; Mehmood, I.; Rho, S. A deep feature-based real-time system for Alzheimer disease stage detection. Multimed. Tools Appl. 2020, 1–19. [Google Scholar] [CrossRef]
- Puente-Castro, A.; Fernandez-Blanco, E.; Pazos, A.; Munteanu, C.R. Automatic assessment of Alzheimer’s disease diagnosis based on deep learning techniques. Comput. Biol. Med. 2020, 120, 103764. [Google Scholar] [CrossRef]
- Islam, J.; Zhang, Y. Brain MRI analysis for Alzheimer’s disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inform. 2018, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bottou, L. On-line Learning and Stochastic Approximations. In On-Line Learning in Neural Networks; Cambridge University Press: Cambridge, UK, 2010; pp. 9–42. [Google Scholar]
- Wen, J.; Thibeau-Sutre, E.; Diaz-Melo, M.; Samper-González, J.; Routier, A.; Bottani, S.; Dormont, D.; Durrleman, S.; Burgos, N.; Colliot, O. Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation. Med. Image Anal. 2020, 63, 101694. [Google Scholar] [CrossRef] [PubMed]
- Hollingshead, A.B. Two Factor Index of Social Position; Yale University Press: New Haven, CT, USA, 1957. [Google Scholar]
- Buckner, R.L.; Head, D.; Parker, J.; Fotenos, A.F.; Marcus, D.; Morris, J.C.; Snyder, A.Z. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: Reliability and validation against manual measurement of total intracranial volume. Neuroimage 2004, 23, 724–738. [Google Scholar] [CrossRef]
- Fotenos, A.F.; Snyder, A.Z.; Girton, L.E.; Morris, J.C.; Buckner, R.L. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology 2005, 64, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Larobina, M.; Murino, L. Medical Image File Formats. J. Digit. Imaging 2014, 27, 200–206. [Google Scholar] [CrossRef]
- Talairach, J.; Tournoux, P. Co-Planar Stereotaxic Atlas of the Human Brain; Thieme: Stuttgart, Germany, 1988. [Google Scholar]
- Styner, M. Parametric estimate of intensity inhomogeneities applied to MRI. IEEE Trans. Med. Imaging 2000, 19, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Suh, C.H.; Shim, W.H.; Kim, S.J.; Roh, J.H.; Lee, J.H.; Kim, M.J.; Park, S.; Jung, W.; Sung, J.; Jahng, G.H. Development and validation of a deep learning-based automatic brain segmentation and classification algorithm for Alzheimer disease using 3D T1-weighted volumetric images. Am. J. Neuroradiol. 2020, 41, 2227–2234. [Google Scholar] [CrossRef]
- Filon, J.R.; Intorcia, A.J.; Sue, L.I.; Vazquez Arreola, E.; Wilson, J.; Davis, K.J.; Sabbagh, M.N.; Belden, C.M.; Caselli, R.J.; Adler, C.H.; et al. Gender differences in Alzheimer disease: Brain atrophy, histopathology burden, and cognition. J. Neuropathol. Exp. Neurol. 2016, 75, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Álvarez-Álvarez, I.; Guillén-Grima, F.; Aguinaga-Ontoso, I. Prevalence and incidence of Alzheimer’s disease in Europe: A meta-analysis. Neurology (Engl. Ed.) 2017, 32, 523–532. [Google Scholar] [CrossRef]
- Whitwell, J.L. The protective role of brain size in Alzheimer’s disease. Expert Rev. Neurother. 2010, 10, 1799–1801. [Google Scholar] [CrossRef] [Green Version]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Tufail, A.B.; Ma, Y.K.; Zhang, Q.N. Binary Classification of Alzheimer’s Disease Using sMRI Imaging Modality and Deep Learning. J. Digit. Imaging 2020, 33, 1073–1090. [Google Scholar] [CrossRef]
- Bereciartua, A.; Picon, A.; Galdran, A.; Iriondo, P. 3D active surfaces for liver segmentation in multisequence MRI images. Comput. Methods Programs Biomed. 2016, 132, 149–160. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE Computer Society: Washington, DC, USA, 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE Computer Society: Washington, DC, USA, 2016; pp. 2818–2826. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, Hawaii, 21–26 July 2016; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2017; pp. 1800–1807. [Google Scholar]
- Picon, A.; Seitz, M.; Alvarez-Gila, A.; Mohnke, P.; Ortiz-Barredo, A.; Echazarra, J. Crop conditional Convolutional Neural Networks for massive multi-crop plant disease classification over cell phone acquired images taken on real field conditions. Comput. Electron. Agric. 2019, 167, 105093. [Google Scholar] [CrossRef]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; Volume 2015, pp. 464–472. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, ICML 2015, Lille, France, 6–11 July 2015; International Machine Learning Society (IMLS): Princeton, NJ, USA, 2015; Volume 1, pp. 448–456. [Google Scholar]
- Heaven, D. Why deep-learning AIs are so easy to fool. Nature 2019, 574, 163–166. [Google Scholar] [CrossRef]
- Kwon, H.; Yoon, H.; Park, K.-W. Multi-Targeted Backdoor: Indentifying Backdoor Attack for Multiple Deep Neural Networks. IEICE Trans. Inf. Syst. 2020, E103.D, 883–887. [Google Scholar] [CrossRef]
- Krcmar, H. Informationsmanagement; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Saltz, J.; Hotz, N.; Wild, D.; Stirling, K. Exploring project management methodologies used within data science teams. In Proceedings of the Americas Conference on Information Systems 2018: Digital Disruption, AMCIS 2018, New Orleans, LA, USA, 16 August 2018; Association for Information Systems: Atlanta, GA, USA, 2018. [Google Scholar]
- Wirth, R.; Wirth, R. CRISP-DM: Towards a standard process model for data mining. In Proceedings of the Fourth International Conference on the Practical Application of Knowledge Discovery and Data Mining, Manchester, UK, 11–13 April 2000; pp. 29–39. [Google Scholar]
- Mcqueen, J.; Meilă, M.; Vanderplas, J.; Zhang, Z. Megaman: Scalable Manifold Learning in Python. J. Mach. Learn. Res. 2016, 17, 1–5. [Google Scholar] [CrossRef]
- Breck, E.; Cai, S.; Nielsen, E.; Salib, M.; Sculley, D. The ML test score: A rubric for ML production readiness and technical debt reduction. In Proceedings of the 2017 IEEE International Conference on Big Data, Big Data 2017, Boston, MA, USA, 11–14 December 2017; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2018; Volume 2018, pp. 1123–1132. [Google Scholar]
- Mehrabi, N.; Morstatter, F.; Saxena, N.; Lerman, K.; Galstyan, A. A Survey on Bias and Fairness in Machine Learning. arXiv 2019, arXiv:1908.09635. [Google Scholar]
- Biessmann, F.; Salinas, D.; Schelter, S.; Schmidt, P.; Lange, D. Deep learning for missing value imputation in tables with non-numerical data. In Proceedings of the International Conference on Information and Knowledge Management, Torino, Italy, 22–26 October 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 2017–2026. [Google Scholar]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.J.; Woolrich, M.W.; Smith, S.M. FSL. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkinson, M. BET2: MR-Based Estimation of Brain, Skull and Scalp Surfaces. In Proceedings of the Eleventh Annual Meeting of the Organization for Human Brain Mapping, Toronto, ON, Canada, 12–16 June 2005. [Google Scholar]


| CDR | OASIS-1 | OASIS-2 | OASIS-2 (Our Subset) |
|---|---|---|---|
| 0 (cognitive normal) | 336 | 206 | 177 |
| 0.5 (very-mild dementia) | 70 | 123 | 98 |
| 1 (mild dementia) | 28 | 41 | 27 |
| 2 (moderate dementia) | 2 | 3 | 3 |
| TOTAL | 436 | 373 | 305 |
| Network | Norm. | Strategies | Input Data | Slices Used | Image Size | Test ACC | Test BAC |
|---|---|---|---|---|---|---|---|
| BrainNet2D | [0, 1] | CLR triangular | 3D | 10 (centered in slice #88) | 224 | 0.81 [0.80, 0.80, 0.80, 0.85, 0.82] | 0.82 [0.81, 0.77, 0.78, 0.86, 0.86] |
| min–max scaling | CLR triangular | 3D | 10 (centered in slice #88) | 224 | 0.81 [0.83, 0.80, 0.80, 0.82, 0.80] | 0.81 [0.80, 0.77, 0.77, 0.88, 0.82] | |
| [0, 1] | CLR triangular Batch Normalization | 3D | 10 (centered in slice #88) | 224 | 0.79 [0.77, 0.78, 0.86, 0.80, 0.72] | 0.79 [0.77, 0.78, 0.86, 0.80, 0.72] | |
| [0, 1] | CLR triangular Sex/Age metadata | 3D | 10 (centered in slice #88) | 224 | 0.79 [0.80, 0.77, 0.77, 0.88, 0.82] | 0.84 [0.83, 0.85, 0.85, 0.81, 0.85] | |
| BrainNet3D | min–max scaling | None | 3D | 176 (all) | 176 | 0.78 [0.77, 0.78, 0.80, 0.78, 0.77] | 0.81 [0.83, 0.77, 0.77, 0.82, 0.85] |
| min–max scaling | Batch Normalization | 3D | 176 (all) | 176 | 0.79 [0.82, 0.80, 0.78, 0.80, 0.77] | 0.83 [0.85, 0.79, 0.82, 0.86, 0.83] | |
| min–max scaling | Batch Normalization Sex/Age metadata | 3D | 176 (all) | 176 | 0.79 [0.78, 0.78, 0.83, 0.80, 0.78] | 0.82 [0.84, 0.77, 0.82, 0.86, 0.81] | |
| min–max scaling | Batch Normalization Sex/Age metadata CLR triangular | 3D | 176 (all) | 176 | 0.80 [0.80, 0.82, 0.85, 0.82, 0.74] | 0.84 [0.88, 0.80, 0.88, 0.88, 0.76] | |
| ResNet18 | [0, 1] | ImageNet weights | 2D | 10 (centered in slice #88) | 224 | 0.78 [0.80, 0.82, 0.77, 0.75, 0.75] | 0.79 [0.85, 0.79, 0.67, 0.80, 0.84] |
| min–max scaling | ImageNet weights | 2D | 10 (centered in slice #88) | 224 | 0.81 [0.80, 0.80, 0.83, 0.80, 0.82] | 0.82 [0.80, 0.80, 0.83, 0.80, 0.82] | |
| min–max scaling | ImageNet weights CLR triangular | 2D | 10 (centered in slice #88) | 224 | 0.81 [0.82, 0.83, 0.86, 0.75, 0.77] | 0.83 [0.82, 0.81, 0.86, 0.80, 0.85] |
| Network | Norm. | Strategies | Input Data | Slices Used | Image Size | Test ACC | Test BAC |
|---|---|---|---|---|---|---|---|
| BrainNet2D | [0, 1] | CLR triangular | 3D | 10 (centered in slice #88) | 224 | 0.92 [0.94, 1.00, 0.92, 0.92, 0.83] | 0.92 [0.93, 1.00, 0.92, 0.92, 0.92] |
| [0, 1] | CLR triangular Sex/Age metadata | 3D | 10 (centered in slice #88) | 224 | 0.82 [0.96, 0.93, 0.92, 0.46, 0.81] | 0.83 [0.96, 0.95, 0.92, 0.50, 0.80] | |
| BrainNet3D | min–max scaling | None | 3D | 176 (all) | 176 | 0.67 [0.68, 0.80, 0.73, 0.51, 0.63] | 0.67 [0.68, 0.81, 0.73, 0.52, 0.64] |
| min–max scaling | Batch Normalization | 3D | 176 (all) | 176 | 0.84 [0.81, 1.00, 0.78, 0.82, 0.79] | 0.84 [0.81, 1.00, 0.78, 0.83, 0.77] | |
| min–max scaling | Sex/Age metadata Batch Normalization | 3D | 176 (all) | 176 | 0.77 [0.70, 0.80, 0.82, 0.77, 0.77] | 0.78 [0.70, 0.81, 0.82, 0.79, 0.77] | |
| min–max scaling | Batch Normalization CLR triangular | 3D | 176 (all) | 176 | 0.79 [0.70, 1.00, 0.69, 0.87, 0.69] | 0.79 [0.70, 1.00, 0.68, 0.88, 0.70] | |
| ResNet18 | [0, 1] | ImageNet weights | 2D | 10 (centered in slice #88) | 224 | 0.92 [0.94, 1.00, 0.94, 0.92, 0.79] | 0.91 [0.94, 1.00, 0.94, 0.92, 0.77] |
| [0, 1] | ImageNet weights CLR triangular | 2D | 10 (centered in slice #88) | 224 | 0.91 [0.94, 0.91, 0.94, 0.95, 0.83] | 0.92 [0.94, 0.94, 0.94, 0.94, 0.84] | |
| min–max scaling | ImageNet weights CLR triangular | 2D | 10 (centered in slice #88) | 224 | 0.93 [0.94, 1.00, 0.96, 0.92, 0.83] | 0.93 [0.94, 1.00, 0.96, 0.92, 0.81] |
| Network | Norm. | Strategies | Input Data | Slices Used | Image Size | Test ACC | Test BAC |
|---|---|---|---|---|---|---|---|
| BrainNet2D | [0, 1] | CLR triangular | 3D | 10 (centered in slice #88) | 224 | 0.88 [0.94, 1.00, 0.78, 0.90, 0.77] | 0.85 [0.94, 1.00, 0.62, 0.92, 0.76] |
| BrainNet3D | min–max scaling | Batch Normalization | 3D | 176 (all) | 176 | 0.77 [0.87, 0.84, 0.80, 0.74, 0.58] | 0.76 [0.87, 0.85, 0.63, 0.82, 0.63] |
| ResNet18 | [0, 1] | ImageNet weights CLR triangular | 2D | 10 (centered in slice #88) | 224 | 0.89 [0.94, 0.98, 0.84, 0.92, 0.79] | 0.88 [0.94, 0.99, 0.67, 0.94, 0.85] |
| CN vs. AD | Multiclass: CN vs. Mild vs. Severe | |||||
|---|---|---|---|---|---|---|
| Method | Approach | Dataset | ACC | BAC | ACC | BAC |
| (PuenteCastro, 2020) [28] | 2D slice level | OASIS-1 | -- | -- | -- | 0.86 |
| (Islam and Zhang, 2018) [29] | 2D slice level (112 × 112 crops) | OASIS-1 | -- | 0.93 | -- | |
| (Wen, 2020) [31] | 2D slice level | OASIS-1 (over 62 years) | -- | 0.68 [0.68, 0.67, 0.69, 0.70, 0.66] | -- | -- |
| 3D subject level | -- | 0.68 [0.65, 0.70, 0.70, 0.71, 0.65] | -- | -- | ||
| Our BrainNet2D | 2D slice level | OASIS-1 | 0.79 [0.80, 0.77, 0.77, 0.88, 0.82] | 0.84 [0.83, 0.85, 0.85, 0.81, 0.85] | ||
| Out BrainNet3D | 3D subject level | OASIS-1 | 0.80 [0.80, 0.82, 0.85, 0.82, 0.74] | 0.84 [0.88, 0.80, 0.88, 0.88, 0.76] | ||
| Our BrainNet2D | 2D slice level | OASIS-2 | 0.82 [0.96, 0.93, 0.92, 0.46, 0.81] | 0.83 [0.96, 0.95 0.92, 0.50, 0.80] | 0.88 [0.94, 1.00, 0.78, 0.90, 0.77] | 0.85 [0.94, 1.00, 0.62, 0.92, 0.76] |
| Out BrainNet3D | 3D subject level | OASIS-2 | 0.84 [0.81, 1.00, 0.78, 0.82, 0.79] | 0.84 [0.81, 1.00, 0.78, 0.83, 0.77] | 0.77 [0.87, 0.84, 0.80, 0.74, 0.58] | 0.76 [0.87, 0.85, 0.63, 0.82, 0.63] |
| ResNet18 | 2D slice level | OASIS-2 | 0.93 [0.94, 1.00, 0.96, 0.92, 0.83] | 0.93 [0.94, 1.00, 0.96, 0.92, 0.81] | 0.89 [0.94, 0.98, 0.84, 0.92, 0.79] | 0.88 [0.94, 0.99, 0.67, 0.94, 0.85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saratxaga, C.L.; Moya, I.; Picón, A.; Acosta, M.; Moreno-Fernandez-de-Leceta, A.; Garrote, E.; Bereciartua-Perez, A. MRI Deep Learning-Based Solution for Alzheimer’s Disease Prediction. J. Pers. Med. 2021, 11, 902. https://doi.org/10.3390/jpm11090902
Saratxaga CL, Moya I, Picón A, Acosta M, Moreno-Fernandez-de-Leceta A, Garrote E, Bereciartua-Perez A. MRI Deep Learning-Based Solution for Alzheimer’s Disease Prediction. Journal of Personalized Medicine. 2021; 11(9):902. https://doi.org/10.3390/jpm11090902
Chicago/Turabian StyleSaratxaga, Cristina L., Iratxe Moya, Artzai Picón, Marina Acosta, Aitor Moreno-Fernandez-de-Leceta, Estibaliz Garrote, and Arantza Bereciartua-Perez. 2021. "MRI Deep Learning-Based Solution for Alzheimer’s Disease Prediction" Journal of Personalized Medicine 11, no. 9: 902. https://doi.org/10.3390/jpm11090902
APA StyleSaratxaga, C. L., Moya, I., Picón, A., Acosta, M., Moreno-Fernandez-de-Leceta, A., Garrote, E., & Bereciartua-Perez, A. (2021). MRI Deep Learning-Based Solution for Alzheimer’s Disease Prediction. Journal of Personalized Medicine, 11(9), 902. https://doi.org/10.3390/jpm11090902