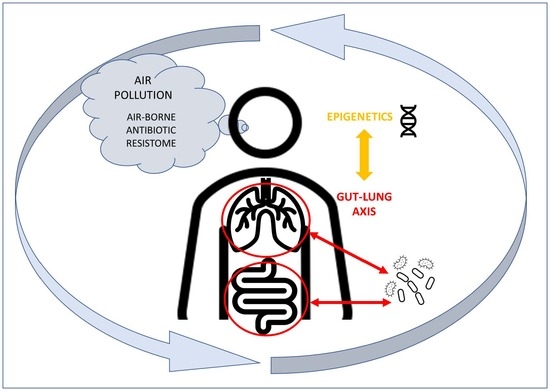
The Interaction among Microbiota, Epigenetic Regulation, and Air Pollutants in Disease Prevention
Abstract
:1. Introduction
2. Early Exposure to Environmental Pollutants, and Dysbiosis as Risk Factor for Late Onset Diseases
The Microbiota and Epigenetic Regulation
3. Air Pollution Can Modify the Intestinal Microbiota
4. The Lung–Gut Axis and the Influence of the Microbiota on the Immune System
5. Metagenomics Approaches to Study Microbial Communities
6. Environmental Antibiotic Pollution and Microbiota: Implication for Public Health
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 30, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.H.; Adcock, I.M.; Chotirmall, S.; Chung, K.F.; et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019, 7, 907–920. [Google Scholar] [CrossRef]
- Hu, J.; Bao, Y.; Zhu, Y.; Osman, R.; Shen, M.; Zhang, Z.; Wang, L.; Cao, S.; Li, L.; Wu, Q. The Preliminary Study on the Association Between PAHs and Air Pollutants and Microbiota Diversity. Arch Environ. Contam. Toxicol 2020, 79, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Adar, S.D.; Huffnagle, G.B.; Curtis, J.L. The respiratory microbiome: An underappreciated player in the human response to inhaled pollutants? Ann Epidemiol. 2016, 26, 355–359. [Google Scholar] [CrossRef] [Green Version]
- Maier, K.L.; Alessandrini, F.; Beck-Speier, I.; Josef Hofer, T.P.; Diabaté, S.; Bitterle, E.; Stöger, T.; Jakob, T.; Behrendt, H.; Horsch, M.; et al. Health effects of ambient particulate matter—Biological mechanisms and inflammatory responses to in vitro and in vivo particle exposures. Inhal. Toxicol. 2008, 20, 319–337. [Google Scholar] [CrossRef] [PubMed]
- Conti, P.; Allegretta, M.; Williams, T.W.; Cifone, M.G.; Alesse, E.; Reale, M.; Boidi, E.; Dempsey, R.A. Enhanced thromboxane synthesis and vacuolization in human polymorphonuclear leucocytes induced by human lymphokine containing supernatants. Clin Rheumatol. 1985, 4, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, X.; Xu, Y.; Wu, W.; Tang, W.; Chen, Z.; Ji, G.; Peng, J.; Jiang, Q.; Xiao, J.; et al. Gut microbiota partially mediates the effects of fine particulate matter on type 2 diabetes: Evidence from a population-based epidemiological study. Environ. Int. 2019, 130, 104882. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Levin, B.J.; Huang, Y.Y.; Peck, S.C.; Wei, Y.; Martínez-Del Campo, A.; Marks, J.A.; Franzosa, E.A.; Huttenhower, C.; Balskus, E.P. A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans-4-hydroxy-l-proline. Science 2017, 355, eaai8386. [Google Scholar] [CrossRef] [Green Version]
- Mah, J.H.; Park, Y.K.; Jin, Y.H.; Lee, J.H.; Hwang, H.J. Bacterial Production and Control of Biogenic Amines in Asian Fermented Soybean Foods. Foods 2019, 8, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elgamal, Z.; Singh, P.; Geraghty, P. The Upper Airway Microbiota, Environmental Exposures, Inflammation, and Disease. Medicina 2021, 57, 823. [Google Scholar] [CrossRef]
- Gruzieva, O.; Xu, C.J.; Yousefi, P.; Relton, C.; Merid, S.K.; Breton, C.V.; Gao, L.; Volk, H.E.; Feinberg, J.I.; Ladd-Acosta, C.; et al. Prenatal Particulate Air Pollution and DNA Methylation in Newborns: An Epigenome-Wide Meta-Analysis. Environ Health Perspect. 2019, 127, 57012. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. mBio 2015, 6, e02288-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Gubory, K.H. Environmental pollutants and lifestyle factors induce oxidative stress and poor prenatal development. Reprod Biomed Online 2014, 29. [Google Scholar] [CrossRef] [Green Version]
- De Steenhuijsen Piters, W.A.; Sanders, E.A.; Bogaert, D. The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef] [Green Version]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015, 17, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.S.; Vestal, G.; Wille, K.; Patel, K.N.; Cheng, F.; Tipparaju, S.; Tousif, S.; Banday, M.M.; Xu, X.; Wilson, L.; et al. Differences in airway microbiome and metabolome of single lung transplant recipients. Respir. Res. 2020, 21, 104. [Google Scholar] [CrossRef]
- Saenen, N.D.; Martens, D.S.; Neven, K.Y.; Alfano, R.; Bové, H.; Janssen, B.G.; Roels, H.A.; Plusquin, M.; Vrijens, K.; Nawrot, T.S. Air pollution-induced placental alterations: An interplay of oxidative stress, epigenetics, and the aging phenotype? Clin. Epigenet. 2019, 11, 124. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Li, S.; Gan, R.Y.; Zhou, T.; Xu, D.P.; Li, H.B. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015, 12, 7493–7519. [Google Scholar] [CrossRef]
- Hugon, P.; Dufour, J.C.; Colson, P.; Fournier, P.E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 10, 1211–1219. [Google Scholar] [CrossRef]
- Alcon-Giner, C.; Caim, S.; Mitra, S.; Ketskemety, J.; Wegmann, U.; Wain, J.; Belteki, G.; Clarke, P.; Hall, L.J. Optimisation of 16S rRNA gut microbiota profiling of extremely low birth weight infants. BMC Genom. 2017, 18, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 44, 15718–15723. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Liu, Y.; Cao, H.; Zhang, Y.; Gu, Z.; Liu, X.; Yu, A.; Kaphle, P.; Dickerson, K.E.; Ni, M.; et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 2020, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.; Raddatz, G.; Gutekunst, J.; Ridnik, M.; Cohen, D.; Abu-Remaileh, M.; Tuganbaev, T.; Shapiro, H.; Pikarsky, E.; Elinav, E.; et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 2020, 5, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Salminen, S.; Verhagen, H.; Rowland, I.; Heimbach, J.; Bañares, S.; Young, T.; Nomoto, K.; Lalonde, M. Novel probiotics and prebiotics: Road to the market. Curr. Opin. Biotechnol. 2015, 32, 99–103. [Google Scholar] [CrossRef]
- Cortese, R.; Lu, L.; Yu, Y.; Ruden, D.; Claud, E.C. Epigenome-Microbiome crosstalk: A potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 2016, 11, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Angrisano, T.; Brancaccio, M.; Falanga, A.; Lombardi, L.; Natale, F.; Laneri, S.; Lombardo, B.; Galdiero, S.; Scudiero, O. Beta-defensins and analogs in Helicobacter pylori infections: mRNA expression levels, DNA methylation, and antibacterial activity. PLoS ONE 2019, 14, e0222295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; de Roos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Kogut, M.H.; Lee, A.; Santin, E. Microbiome and pathogen interaction with the immune system. Poult. Sci. 2020, 99, 1906–1913. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Hauth, J.C.; Andrews, W.W. Intrauterine infection and preterm delivery. N. Engl. J. Med. 2000, 20, 1500–1507. [Google Scholar] [CrossRef]
- Guamer, F. Role of intestinal flora in health and disease. Nutr. Hosp. 2007, 22, 14–19. [Google Scholar]
- Sekirov, I.; Gill, N.; Jogova, M.; Tam, N.; Robertson, M.; de Llanos, R.; Li, Y.; Finlay, B.B. Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 2010, 1, 30–41. [Google Scholar] [CrossRef] [Green Version]
- Mutlu, E.A.; Engen, P.A.; Soberanes, S. Particulate matter air pollution causes oxidant-mediated increase in gut permeability in mice. Part. Fibre Toxicol. 2011, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouladi, F.; Bailey, M.J.; Patterson, W.B.; Sioda, M.; Blakley, I.C.; Fodor, A.A.; Jones, R.B.; Chen, Z.; Kim, J.S.; Lurmann, F.; et al. Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ. Int. 2020, 138, 105604. [Google Scholar] [CrossRef]
- Alderete, T.L.; Jones, R.B.; Chen, Z.; Kim, J.S.; Habre, R.; Lurmann, F.; Gilliland, F.D.; Goran, M.I. Exposure to traffic-related air pollution and the composition of the gut microbiota in overweight and obese adolescents. Environ. Res. 2018, 161, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Beamish, L.A.; Osornio-Vargas, A.R.; Wine, E.J. Air pollution: An environmental factor contributing to intestinal disease. Crohns Colitis 2011, 5, 279–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elder, A.; Oberdörster, G. Translocation and effects of ultrafine particles outside of the lung. Clin. Occup. Environ. Med. 2006, 5, 785–796. [Google Scholar]
- Salim, S.Y.; Kaplan, G.G.; Madsen, K.L. Air pollution effects on the gut microbiota: A link between exposure and inflammatory disease. Gut Microbes 2014, 5, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Rajani, C.A. The Influence of Gut Microbial Metabolism on the Development and Progression of Non-alcoholic Fatty Liver Disease. Exp. Med. Biol. 2018, 1061, 95–110. [Google Scholar]
- Li, K.J.; Chen, Z.L.; Huang, Y.; Zhang, R.; Luan, X.Q.; Lei, T.T. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir Res. 2019, 20, 272. [Google Scholar] [CrossRef]
- Yasuyuki, M.; Kunihiro, K.; Kurissery, S.; Kanavillil, N.; Sato, Y.; Kikuchi, Y. Antibacterial properties of nine pure metals: A laboratory study using Staphylococcus aureus and Escherichia coli. Biofouling 2010, 26, 851–858. [Google Scholar] [CrossRef]
- Gackière, F.; Saliba, L.; Baude, A.; Bosler, O.; Strube, C. Ozone inhalation activates stress-responsive regions of the CNS. J. Neurochem. 2011, 117, 961–972. [Google Scholar] [CrossRef]
- Thomson, E.M.J. Air Pollution, Stress, and Allostatic Load: Linking Systemic and Central Nervous System Impacts. Alzheimers Dis. 2019, 69, 597–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, G.G.; Hubbard, J.; Korzenik, J.; Sands, B.E.; Panaccione, R.; Ghosh, S. The inflammatory bowel diseases and ambient air pollution: A novel association. Am. J. Gastroenterol. 2010, 105, 2412–2419. [Google Scholar] [CrossRef] [Green Version]
- López-Abente, G.; García-Pérez, J.; Fernández-Navarro, P.; Boldo, E.; Ramis, R. Colorectal cancer mortality and industrial pollution in Spain. BMC Public Health 2012, 12, 589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutlu, E.A.; Comba, I.Y.; Cho, T.; Engen, P.A.; Yazıcı, C.; Soberanes, S.; Hamanaka, R.B.; Niğdelioğlu, R.; Meliton, A.Y.; Ghio, A.J.; et al. Inhalational exposure to particulate matter air pollution alters the composition of the gut microbiome. Environ. Pollut. 2018, 240, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; An, Y.; Zhou, J.; Yang, J.; Zhang, Y.; Yang, J.; Wang, L.; Li, X.; Lu, D.; Zhon, J.; et al. Subchronic exposure to concentrated ambient PM2.5 perturbs gut and lung microbiota as well as metabolic profiles in mice. Environ. Pollut. 2021, 272, 115987. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, T.; Si, B.; Du, H.; Liu, Y.; Waqas, A.; Huang, S.; Zhao, G.; Chen, S.; Xu, A. Intratracheally instillated diesel PM2.5 significantly altered the structure and composition of indigenous murine gut microbiota. Ecotoxicol. Environ. Saf. 2021, 210, 111903. [Google Scholar] [CrossRef]
- Curciarello, R.; Canziani, K.E.; Docena, G.H.; Muglia, C.I. Contribution of Non-immune Cells to Activation and Modulation of the Intestinal Inflammation. Front Immunol. 2019, 10, 647. [Google Scholar] [CrossRef] [Green Version]
- Bailey, J.M.; Noopur, N.; Naik, N.N.; Laura, E.; Wild, E.L.; Patterson, B.W.; Alderete, L.T. Exposure to air pollutants and the gut microbiota: A potential link between exposure, obesity, and type 2 diabetes. Gut Microbes 2020, 11, 1188–1202. [Google Scholar] [CrossRef]
- Vari, H.K.; Roslund, M.I.; Oikarinen, S.; Nurminen, N.; Puhakka, R.; Parajuli, A.; Grönroos, M.; Siter, N.; Laitinen, O.H.; Hyöty, H.; et al. ADELE research group. Associations between land cover categories, gaseous PAH levels in ambient air and endocrine signaling predicted from gut bacterial metagenome of the elderly. Chemosphere 2021, 265, 128965. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Kwon, D.I.; Kim, M.; Im, S.H.; Lee, Y.J. Commensal Microbiome Expands Tγδ17 Cells in the Lung and Promotes Particulate Matter-Induced Acute Neutrophilia. Front Immunol. 2021, 12, 645741. [Google Scholar] [CrossRef]
- Yang, K. Ultrafine particles altered gut microbial population and metabolic profiles in a sex-specific manner in an obese mouse. Sci. Rep. 2021, 25, 6906. [Google Scholar] [CrossRef]
- Van den Brule, S.; Rappe, M.; Ambroise, J.; Bouzin, C.; Dessy, C.; Paquot, A.; Muccioli, G.G.; Lison, D. Diesel exhaust particles alter the profile and function of the gut microbiota upon subchronic oral administration in mice. Part. Fibre Toxicol. 2021, 18, 7. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.M.; Nordgren, M.T.; McCole, F.D. Every breath you take: Impacts of environmental dust exposure on intestinal barrier function—From the gut-lung axis to COVID-19. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G586–G600. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Maruvada, P.; Leone, V.; Kaplan, L.M.; Chang, E.B. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe 2017, 22, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Lejeune, A.; Scher, G.C.J.; Koralov, S.B. Microbial-derived antigens and metabolites in spondyloarthritis. Semin. Immunopathol. 2021, 43, 163–172. [Google Scholar] [CrossRef]
- Woodby, B.; Schiavone, M.L.; Pambianchi, E.; Mastaloudis, A.; Hester, S.N.; Wood, S.M.; Pecorelli, A.; Valacchi, A. Particulate matter decreases intestinal barrier-associated proteins levels in 3D human intestinal model. Int. J. Environ. Res. Public Health 2020, 17, 3234. [Google Scholar] [CrossRef]
- Alemao, C.A.; Budden, K.F.; Gomez, H.M.; Rehman, S.F.; Marshall, J.E.; Shukla, S.D.; Donovan, C.; Forster, S.C.; Yang, I.A.; Keely, S.; et al. Impact of diet and the bacterial microbiome on the mucous barrier and immune disorders. Allergy 2021, 76, 714–734. [Google Scholar] [CrossRef]
- Xue, Y.; Chu, J.; Li, Y.; Kong, X. The influence of air pollution on respiratory microbiome: A link to respiratory disease. Toxicol. Lett. 2020, 334, 14–20. [Google Scholar] [CrossRef]
- Mariani, J.; Favero, C.; Spinazzè, A.; Cavallo, D.M.; Carugno, M.; Motta, V.; Bonzini, M.; Cattaneo, A.; Pesatori, A.C.; Bollati, V. Short-term particulate matter exposure influences nasal microbiota in a population of healthy subjects. Environ. Res. 2018, 162, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Vignal, C.; Guilloteau, E.; Gower-Rousseau, C.; Body-Malapel, M. Review article: Epidemiological and animal evidence for the role of air pollution in intestinal diseases. Sci. Total Environ. 2021, 757, 143718. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Mohanty, D. Psychobiotics: A new approach for treating mental illness? Crit. Rev. Food Sci. Nutr. 2019, 59, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef]
- Chu, C.; Murdock, M.H.; Jing, D.; Won, T.H.; Chung, H.; Kressel, A.M.; Tsaava, T.; Addorisio, M.E.; Putzel, G.G.; Zhou, L.; et al. The microbiota regulate neuronal function and fear extinction learning. Nature 2019, 574, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Siljander, H.; Honkanen, J.; Knip, M. Microbiome and type 1 diabetes. EBioMedicine 2019, 46, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Zununi Vahed, S.; Moghaddas Sani, H.; Rahbar Saadat, Y.; Barzegari, A.; Omidi, Y. Type 1 diabetes: Through the lens of human genome and metagenome interplay. Biomed. Pharmacother. 2018, 104, 332–342. [Google Scholar] [CrossRef]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung–gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal. Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Brunekreef, B.; Downward, G.; Forastiere, F.; Gehring, U.; Heederik, D.J.J.; Hoek, G.; Koopmans, M.P.G.; Smith, L.A.M.; Vermeulen, R.C.H. Air Pollution and COVID-19. Eur. Parliam. 2021, 2, 317–328. [Google Scholar]
- Franklin, B.A.; Brook, R.; Arden Pope, C., III. Air pollution and cardiovascular disease. Curr. Probl. Cardiol. 2015, 40, 207–238. [Google Scholar] [CrossRef]
- Hamra, G.B.; Guha, N.; Cohen, A.; Laden, F.; Raaschou-Nielsen, O.; Samet, J.M. Outdoor particulate matter exposure and lung cancer. Environ. Health Perspect. 2014, 122, 906–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreyling, W.G.; Semmler, M.; Möller, W. Dosimetry and toxicology of ultrafine particles. J. Aerosol Med. 2004, 17, 140–152. [Google Scholar] [CrossRef]
- Kreyling, W.G.; Dirscherl, P.; Ferron, G.A.; Heilmann, P.; Josten, M.; Miaskowski, U.; DNeuner, M.; Reitmeir, P.; Ruprecht, L.; Schumann, G.; et al. Health effects of sulfur-related environmental air pollution. III. Nonspecific respiratory defense capacities. Inhal Toxicol. 1999, 11, 391–422. [Google Scholar]
- Semmler-Behnke, M.; Takenaka, S.; Fertsch, S.; Wenk, A.; Seitz, J.; Mayer, P.; Oberdörster, G.; Kreyling, W.G. Efficient elimination of inhaled nanoparticles from the alveolar region: Evidence for interstitial uptake and subsequent reentrainment onto airways epithelium. Environ. Health Perspect. 2007, 115, 728–733. [Google Scholar] [CrossRef] [Green Version]
- De Brouwere, K.; Buekers, J.; Cornelis, C.; Schlekat, C.E.; Oller, A.R. Assessment of indirect human exposure to environmental sources of nickel: Oral exposure and risk characterization for systemic effects. Sci. Total Environ. 2012, 419, 25–36. [Google Scholar] [CrossRef]
- Tilly-Kiesi, M.; Schaefer, E.J.; Knudsen, P.; Welty, F.K.; Dolnikowski, G.G.; Taskinen, M.R.; Lichtenstein, A.H. Lipoprotein metabolism in sujects with hepatic lipase deficiency. Metabolism 2004, 53, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.L.; Hedin, C.R.H.; Koutsoumpas, A.; Ng, S.C.; McCarthy, N.E.; Prescott, N.J.; Pessoa-Lopes, P.; Mathew, C.G.; Sanderson, J.; Hart, A.L.; et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm. Bowel Dis. 2012, 18, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Birchenough, G.M.H.; Ståhlman, M.; Arike, L.; Johansson, M.E.V.; Hansson, G.C.; Bäckhed, F. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 2018, 23, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrakhovitch, E.A.; Tabibzadeh, S. Homocysteine in Chronic Kidney Disease. Adv. Clin. Chem. 2015, 72, 77–106. [Google Scholar] [PubMed]
- Ringh, M.V.; Hagemann-Jensen, M.; Needhamsen, M.; Kular, L.; Breeze, C.E.; Sjöholm, L.K.; Slavec, L.; Kullberg, S.; Wahlström, J.; Grunewald, J.; et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. BioMedicine 2019, 46, 290–304. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.R.; Chida, A.S.; Bauter, M.R.; Shafiq, N.; Seweryniak, K.; Maggirwar, S.B.; Kilty, I.; Rahman, I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am. J. Physiol. Lung Cell Mol. Physiol. 2006, 291, L46–L57. [Google Scholar] [CrossRef] [Green Version]
- The Committee on Metagenomics. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet; The National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Li, M.; Wen, J. Recent progress in the application of omics technologies in the study of bio-mining microorganisms from extreme environments. Microb. Cell Fact. 2021, 20, 178. [Google Scholar] [CrossRef]
- Tringe, S.G.; von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [Green Version]
- Gaur, V.K.; Gupta, S.; Pandey, A. Evolution in mitigation approaches for petroleum oil-polluted environment: Recent advances and future directions. Environ. Sci. Pollut. Res. Int. 2021. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Wani, G.A.; Khan, M.A.; Dar, M.A.; Shah, M.A.; Reshi, Z.A. Next Generation High Throughput Sequencing to Assess Microbial Communities: An Application Based on Water Quality. Bull. Environ. Contam. Toxicol. 2021, 106, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Young, R.B.; Marcelino, V.R.; Chonwerawong, M.; Gulliver, E.L.; Forster, S.C. Key Technologies for Progressing Discovery of Microbiome-Based Medicines. Front. Microbiol. 2021, 12, 685935. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 29 September 2021).
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Gillings, M.; Simonet, P.; Stekel, D.; Banwart, S.; Penuelas, J. Microbial mass movements. Science 2017, 357, 1099–1100. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Jin, L.; He, T.; Chen, B.; Luo, X.; Feng, B.; Huang, W.; Li, J.; Fu, P.; Li, X. Bacteria and Antibiotic Resistance Genes (ARGs) in PM2.5 from China: Implications for Human Exposure. Environ. Sci. Technol. 2019, 52, 963–972. [Google Scholar] [CrossRef]
- Danko, D.; Bezdan, D.; Afshin, E.E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; Donnellan, D.; Hecht, J.; Jackson, K.; et al. International MetaSUB Consortium. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell 2021, 184, 3376–3393.e17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, J.; Zhu, Y.G.; Chen, Q.L.; Shen, F.; Wu, Y.; Xu, S.; Fan, H.; Da, G.; Huang, R.J.; et al. Global Survey of Antibiotic Resistance Genes in Air. Environ. Sci. Technol. 2018, 52, 10975–10984. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Zhao, F.; Zhang, X.X.; Li, K.; Li, C.; Ye, L.; Li, M. Metagenomic profiling of ARGs in airborne particulate matters during a severe smog event. Sci. Total Environ. 2018, 615, 1332–1340. [Google Scholar] [CrossRef]
- He, P.; Wu, Y.; Huang, W.; Wu, X.; Lv, J.; Liu, P.; Bu, L.; Bai, Z.; Chen, S.; Feng, W.; et al. Characteristics of and variation in airborne ARGs among urban hospitals and adjacent urban and suburban communities: A metagenomic approach. Environ. Int. 2020, 139, 105625. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The gut-lung axis in health and respiratory diseases: A place for inter-organ and inter-kingdom crosstalks. Front. Cell Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Liu, Y.; Li, S.; Peng, Z.; Liu, X.; Chen, J.; Zheng, X. Role of lung and gut microbiota on lung cancer pathogenesis. J. Cancer Res. Clin. Oncol. 2021, 14, 2177–2186. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Particulate Matter | Microbiota | References |
|---|---|---|
| PM2.5 exposure in mice | Lung/intestinal damage and systemic inflammatory reactions | [51] |
| Inhaled diesel PM2.5 in mice | Alteration of gut microbiota diversity and community | [7] |
| PM can be indirectly deposited in oropharynx via mucociliary clearance and upon swallowing of saliva and mucus | Alteration of the GI epithelium and gut microbiome | [49] |
| Antibiotics, air pollutants, lifestyle, diet, breast feeding | Mucosal inflammation | [52] |
| Particulate matter, nitrogen oxides, and ozone | Alteration of the gut microbiota with risk of obesity and type 2 diabetes | [53] |
| Traffic-related air pollution | Gut microbial taxa and fasting glucose levels | [38] |
| Polycyclic aromatic hydrocarbons (PAHs) | Modulation of endocrine signaling pathways in gut microbiota | [54] |
| Particulate matter (PM) | PM-induced neutrophilia | [55] |
| Air pollution | Increased risk of metabolic dysfunction in obese individuals | [56] |
| Particulate matter including diesel exhaust particles | At relevant doses, changes the composition and function of the gut microbiota | [57] |
| Particulate matter | Promote Pseudomonas aeruginosa infection | [50] |
| Particulate matter | multiple gastrointestinal symptoms in patients with COVID-19 and progression with special emphasis on the lung–gut axis | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pulliero, A.; Traversi, D.; Franchitti, E.; Barchitta, M.; Izzotti, A.; Agodi, A. The Interaction among Microbiota, Epigenetic Regulation, and Air Pollutants in Disease Prevention. J. Pers. Med. 2022, 12, 14. https://doi.org/10.3390/jpm12010014
Pulliero A, Traversi D, Franchitti E, Barchitta M, Izzotti A, Agodi A. The Interaction among Microbiota, Epigenetic Regulation, and Air Pollutants in Disease Prevention. Journal of Personalized Medicine. 2022; 12(1):14. https://doi.org/10.3390/jpm12010014
Chicago/Turabian StylePulliero, Alessandra, Deborah Traversi, Elena Franchitti, Martina Barchitta, Alberto Izzotti, and Antonella Agodi. 2022. "The Interaction among Microbiota, Epigenetic Regulation, and Air Pollutants in Disease Prevention" Journal of Personalized Medicine 12, no. 1: 14. https://doi.org/10.3390/jpm12010014
APA StylePulliero, A., Traversi, D., Franchitti, E., Barchitta, M., Izzotti, A., & Agodi, A. (2022). The Interaction among Microbiota, Epigenetic Regulation, and Air Pollutants in Disease Prevention. Journal of Personalized Medicine, 12(1), 14. https://doi.org/10.3390/jpm12010014