GSTO1, GSTO2 and ACE2 Polymorphisms Modify Susceptibility to Developing COVID-19
Abstract
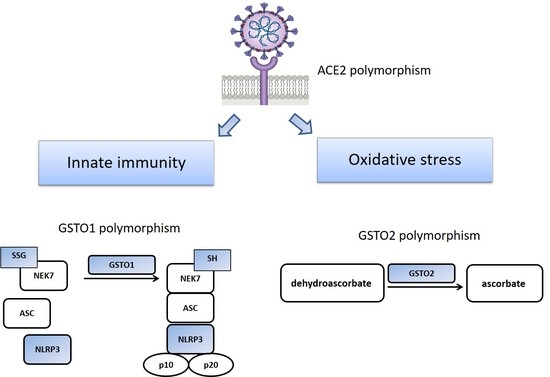
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lavillette, D.; Barbouche, R.; Yao, Y.; Boson, B.; Cosset, F.-L.; Jones, I.M.; Fenouillet, E. Significant redox insensitivity of the functions of the SARS-CoV spike glycoprotein. J. Biol. Chem. 2006, 281, 9200–9204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Board, P.G.; Coggan, M.; Chelvanayagam, G.; Easteal, S.; Jermiin, L.S.; Schulte, G.K.; Danley, D.E.; Hoth, L.R.; Griffor, M.C.; Kamath, A.V.; et al. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 2000, 275, 24798–24806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Board, P.G. The omega-class glutathione transferases: Structure, function, and genetics. Drug Metab. Rev. 2011, 43, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Whitbread, A.K.; Tetlow, N.; Eyre, H.J.; Sutherland, G.R.; Board, P.G. Characterization of the human omega class glutathione transferase genes and associated polymorphisms. Pharmacogenetics 2003, 13, 131–144. [Google Scholar] [CrossRef]
- Harju, T.H.; Peltoniemi, M.J.; Rytilä, P.H.; Soini, Y.; Salmenkivi, K.M.; Board, P.G.; Ruddock, L.W.; Kinnula, V.L. Glutathione S-transferase omega in the lung and sputum supernatants of COPD patients. Respir. Res. 2007, 8, 48. [Google Scholar] [CrossRef] [Green Version]
- Menon, D.; Coll, R.; O’Neill, L.A.J.; Board, P.G. GSTO1-1 modulates metabolism in macrophages activated through the LPS and TLR4 pathway. J. Cell Sci. 2015, 128, 1982–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, D.; Innes, A.; Oakley, A.J.; Dahlstrom, J.E.; Jensen, L.M.; Brüstle, A.; Tummala, P.; Rooke, M.; Casarotto, M.G.; Baell, J.B.; et al. GSTO1-1 plays a pro-inflammatory role in models of inflammation, colitis and obesity. Sci. Rep. 2017, 7, 17832. [Google Scholar] [CrossRef] [PubMed]
- Majidi, N.; Rabbani, F.; Gholami, S.; Gholamalizadeh, M.; BourBour, F.; Rastgoo, S.; Hajipour, A.; Shadnoosh, M.; Akbari, M.E.; Bahar, B.; et al. The effect of Vitamin C on pathological parameters and survival duration of critically ill coronavirus disease 2019 patients: A randomized clinical trial. Front. Immunol. 2021, 12, 717816. [Google Scholar] [CrossRef] [PubMed]
- Wilk, J.B.; Walter, R.E.; Laramie, J.M.; Gottlieb, D.J.; O’Connor, G.T. Framingham heart study genome-wide association: Results for pulmonary function measures. BMC Med. Genet. 2007, 8, S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-H.; Yeh, S.-D.; Shen, K.-H.; Shen, C.-H.; Juang, G.-D.; Hsu, L.-I.; Chiou, H.-Y.; Chen, C.-J. A significantly joint effect between arsenic and occupational exposures and risk genotypes/diplotypes of CYP2E1, GSTO1 and GSTO2 on risk of urothelial carcinoma. Toxicol. Appl. Pharmacol. 2009, 241, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Board, P.G.; Menon, D. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 3267–3288. [Google Scholar] [CrossRef] [PubMed]
- Tanaka-Kagawa, T.; Jinno, H.; Hasegawa, T.; Makino, Y.; Seko, Y.; Hanioka, N.; Ando, M. Functional characterization of two variant human GSTO 1-1s (Ala140Asp and Thr217Asn). Biochem. Biophys. Res. Commun. 2003, 301, 516–520. [Google Scholar] [CrossRef]
- Menon, D.; Board, P.G. A role for glutathione transferase Omega 1 (GSTO1-1) in the glutathionylation cycle. J. Biol. Chem. 2013, 288, 25769–25779. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.; Zou, F.; Chai, H.S.; Younkin, C.S.; Miles, R.; Nair, A.A.; Crook, J.E.; Pankratz, V.S.; Carrasquillo, M.M.; Rowley, C.N.; et al. Glutathione S-transferase omega genes in alzheimer and parkinson disease risk, age-at-diagnosis and brain gene expression: An Association Study with Mechanistic Implications. Mol. Neurodegener. 2012, 7, 13. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, B.; Salavaggione, O.E.; Pelleymounter, L.L.; Moon, I.; Eckloff, B.W.; Schaid, D.J.; Wieben, E.D.; Weinshilboum, R.M. Glutathione S-transferase omega 1 and omega 2 pharmacogenomics. Drug Metab. Dispos. 2006, 34, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.S. TMPRSS2: An equally important protease as ACE2 in the pathogenicity of SARS-CoV-2 infection. Mayo Clin. Proc. 2021, 96, 2748–2752. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhu, Y.; Lan, J.; Chen, H.; Wang, Y.; Shi, H.; Feng, F.; Chen, D.-Y.; Close, B.; Zhao, X.; et al. Susceptibilities of human ACE2 genetic variants in coronavirus infection. J. Virol. 2022, 96, e01492-21. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Xu, X.; Zhou, X.; Chen, P.; Liang, H.; Li, X.; Zhong, W.; Hao, P. Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. J. Gen. Virol. 2020, 101, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.T.; Evans, D.M.; An, J.; Jones, S.J.M. ACE 2 coding variants: A potential X-linked risk factor for COVID-19 disease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Miljanovic, D.; Milicevic, O.; Loncar, A.; Abazovic, D.; Despot, D.; Banko, A. The first molecular characterization of Serbian SARS-CoV-2 isolates from a unique early second wave in Europe. Front. Microbiol. 2021, 12, 1526. [Google Scholar] [CrossRef] [PubMed]
- Musaogullari, A.; Chai, Y.-C. Redox regulation by protein S-glutathionylation: From molecular mechanisms to implications in health and disease. Int. J. Mol. Sci. 2020, 21, 8113. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.M.; Hooftman, A.; Angiari, S.; Tummala, P.; Zaslona, Z.; Runtsch, M.C.; McGettrick, A.F.; Sutton, C.E.; Diskin, C.; Rooke, M.; et al. Glutathione transferase omega-1 regulates NLRP3 inflammasome activation through NEK7 deglutathionylation. Cell Rep. 2019, 29, 151–161.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, N.; Di, B.; Xu, L. The NLRP3 inflammasome and COVID-19: Activation, pathogenesis and therapeutic strategies. Cytokine Growth Factor Rev. 2021, 61, 2–15. [Google Scholar] [CrossRef] [PubMed]
- McKee, C.M.; Coll, R.C. NLRP3 inflammasome priming: A riddle wrapped in a mystery inside an enigma. J. Leukoc. Biol. 2020, 108, 937–952. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.S.; de Sá, K.S.G.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021, 218, e20201707. [Google Scholar] [CrossRef] [PubMed]
- Piaggi, S.; Marchi, E.; Carnicelli, V.; Zucchi, R.; Griese, M.; Hector, A.; Sorio, C.; Pompella, A.; Corti, A. Airways glutathione S-transferase omega-1 and its A140D polymorphism are associated with severity of inflammation and respiratory dysfunction in cystic fibrosis. J. Cyst. Fibros. 2021, 20, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, A.; Riazalhosseini, Y. Epigenome aberrations: Emerging driving factors of the clear cell renal cell carcinoma. Int. J. Mol. Sci. 2017, 18, 1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Schellhorn, H.E. New developments and novel therapeutic perspectives for Vitamin C. J. Nutr. 2007, 137, 2171–2184. [Google Scholar]
- Badraoui, R.; Adnan, M.; Bardakci, F.; Alreshidi, M.M. Chloroquine and hydroxychloroquine interact differently with ACE2 domains reported to bind with the coronavirus spike protein: Mediation by ACE2 polymorphism. Molecules 2021, 26, 673. [Google Scholar] [CrossRef]
- Yi, M.C.; Khosla, C. Thiol–disulfide exchange reactions in the mammalian extracellular environment. Annu. Rev. Chem. Biomol. Eng. 2016, 7, 197–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hati, S.; Bhattacharyya, S. Impact of thiol–disulfide balance on the binding of COVID-19 spike protein with angiotensin-converting enzyme 2 receptor. ACS Omega 2020, 5, 16292–16298. [Google Scholar] [CrossRef] [PubMed]
| Baseline Characteristic | COVID-19 Patients (n = 255) | Controls (n = 236) | OR (95%CI) | p |
|---|---|---|---|---|
| Age (years) a | 52.00 ± 12.67 | 50.00 ± 14.14 | / | 0.100 |
| Gender, n (%) | ||||
| Male | 139 (54.5) | 127 (53.8) | 1.00 b | |
| Female | 116 (45.5) | 109 (46.2) | 0.97 (0.68–1.39) | 0.877 |
| Hypertension, n (%) c | ||||
| No | 98 (54.1) | 152 (71.7) | 1.00 b | |
| Yes | 83 (45.9) | 60 (28.3) | 2.15 (1.41–3.26) | <0.001 |
| Obesity, n (%) c | ||||
| BMI < 30 | 159 (64.4) | 161 (83.9) | 1.00 b | |
| BMI > 30 | 88 (35.6) | 31 (16.1) | 2.87 (1.81–4.57) | <0.001 |
| BMI (kg/m2) a | 28.82 ± 5.25 | 26.09 ± 4.29 | / | <0.001 |
| Smoking, n (%) c | ||||
| Never | 133 (54.5) | 88 (37.8) | 1.00 b | |
| Former | 73 (29.9) | 31 (13.3) | 1.56 (0.95–2.56) | 0.082 |
| Ever | 38 (15.6) | 114 (48.9) | 0.22 (0.14–0.35) | <0.001 |
| Diabetes, n (%) c | ||||
| No | 228 (89.4) | 223 (94.5) | 1.00 b | |
| Yes | 27 (10.6) | 13 (5.5) | 2.03 (1.02–4.04) | 0.043 |
| Biomarkers a | Mild COVID-19 (n = 82) | Severe COVID-19 (n = 169) | Reference Values | p |
|---|---|---|---|---|
| Hematologic | ||||
| WBC count (109/L) | 5.62 ± 2.30 | 6.21 ± 2.54 | 3.4–9.7 | 0.121 |
| Lymphocyte count (109/L) | 1.61 ± 0.62 | 1.35 ± 1.34 | 1.2–3.4 | <0.001 |
| Monocyte count (109/L) | 0.62 ± 0.45 | 0.48 ± 0.28 | 0.10–0.80 | 0.019 |
| Platelet count (109/L) | 219.98 ± 59.25 | 205.15 ± 77.46 | 158–424 | 0.033 |
| RBC count (1012/L) | 4.81 ± 0.44 | 4.70 ± 0.54 | 3.86–5.08 | 0.213 |
| Hemoglobin (g/L) | 141.56 ± 12.55 | 135.55 ± 23.75 | 119–157 | 0.063 |
| Biochemical | ||||
| ALT (U/L) | 45.54 ± 31.44 | 53.67 ± 34.04 | 0–41 | 0.073 |
| AST (U/L) | 28.00 ± 15.40 | 41.30 ± 24.36 | 0–37 | <0.001 |
| LDH (U/L) | 212.78 ± 113.83 | 301.89 ± 193.77 | 220–460 | <0.001 |
| Urea (mmol/L) | 4.83 ± 1.78 | 6.17 ± 4.86 | 2.5–7.5 | 0.005 |
| Creatinine (µmol/L) | 77.95 ± 16.65 | 92.80 ± 47.15 | 45–84 | 0.009 |
| Fe (µmol/L) | 10.31 ± 6.43 | 6.07 ± 4.05 | 7.0–28.0 | 0.002 |
| TIBC (µmol/L) | 61.56 ± 63.84 | 43.25 ± 10.57 | 44.8–75.1 | 0.039 |
| Coagulation | ||||
| D-dimer (mg/L) | 0.57 ± 0.43 | 1.36 ± 6.44 | <0.5 | 0.046 |
| Inflammatory biomarkers | ||||
| CRP (mg/L) | 15.36 ± 34.33 | 65.22 ± 64.05 | 0.0–5.0 | <0.001 |
| Fibrinogen (g/L) | 3.10 ± 0.86 | 4.32 ± 1.46 | 2.0–4.0 | <0.001 |
| Serum ferritin (µg/L) | 293.94 ± 394.02 | 707.39 ± 740.24 | 13.0–150.0 | <0.001 |
| IL-6 (pg/mL) | 9.90 ± 17.78 | 40.93 ± 44.28 | <7 | <0.001 |
| Genotype | COVID-19 Patients n, % | Controls n, % | Adjusted OR (95%CI) a | p |
|---|---|---|---|---|
| GSTO1 (rs4925) | ||||
| CC (AlaAla) | 132 (52.4) | 126 (54.3) | 1.00 b | |
| CA (AlaAsp) | 78 (31.0) | 88 (37.9) | 1.42 (0.83–2.42) | 0.205 |
| AA (AlaAsp) | 42 (16.7) | 18 (7.8) | 2.45 (1.03–5.84) | 0.044 |
| GSTO2 (rs156697) | ||||
| AA (AsnAsn) | 115 (45.1) | 137 (60.1) | 1.00 b | |
| AG (AsnAsp) | 89 (34.9) | 74 (32.5) | 1.91 (1.10–3.30) | 0.020 |
| GG (AspAsp) | 51 (20.0) | 17 (7.5) | 3.69 (1.62–8.40) | 0.002 |
| ACE2 (rs4646116) | ||||
| TT (LysLys) | 195 (77.7) | 179 (89.5) | 1.00 b | |
| TC + CC (LysArg + ArgArg) | 56 (22.3) | 21 (10.5) | 1.79 (0.89–3.57) | 0.100 |
| Haplotype | GSTO1 rs4925 | GSTO2 rs156697 | Controls % | COVID 19 Patients % | Adjusted OR (95%CI) a | p |
|---|---|---|---|---|---|---|
| H1 | *C | *A | 66.13 | 56.37 | 1.00 b | |
| H2 | *A | *G | 16.75 | 26.18 | 1.97 (1.28–3.03) | 0.002 |
| H3 | *C | *G | 7.15 | 11.28 | 1.95 (1.00–3.81) | 0.050 |
| H4 | *A | *A | 9.97 | 6.18 | 0.97 (0.48–1.95) | 0.920 |
| Number of Risk-Associated Genotypes | COVID-19 Patients n (%) | Controls n (%) | OR (95%CI) a | p |
|---|---|---|---|---|
| 0 | 72 (29.0) | 82 (41.4) | 1 b | |
| 1 | 67 (27.0) | 61 (30.8) | 1.45 (0.77–2.73) | 0.248 |
| 2 | 86 (34.7) | 50 (25.3) | 2.50 (1.34–4.64) | 0.004 |
| 3 | 23 (9.3) | 5 (2.5) | 3.03 (0.94–9.78) | 0.064 |
| Number of Risk-Associated Genotypes | Mild COVID-19 Patients n (%) | Severe COVID-19 Patients n (%) | OR (95%CI) a | p |
|---|---|---|---|---|
| 0 | 19 (24.1) | 53 (29.5) | 1 b | |
| 1 | 21 (26.6) | 44 (26.6) | 0.58 (0.18–1.92) | 0.371 |
| 2 | 31 (39.2) | 53 (34.4) | 1.07(0.37–3.06) | 0.906 |
| 3 | 8 (10.1) | 15 (9.4) | 0.93 (0.19–4.50) | 0.926 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djukic, T.; Stevanovic, G.; Coric, V.; Bukumiric, Z.; Pljesa-Ercegovac, M.; Matic, M.; Jerotic, D.; Todorovic, N.; Asanin, M.; Ercegovac, M.; et al. GSTO1, GSTO2 and ACE2 Polymorphisms Modify Susceptibility to Developing COVID-19. J. Pers. Med. 2022, 12, 458. https://doi.org/10.3390/jpm12030458
Djukic T, Stevanovic G, Coric V, Bukumiric Z, Pljesa-Ercegovac M, Matic M, Jerotic D, Todorovic N, Asanin M, Ercegovac M, et al. GSTO1, GSTO2 and ACE2 Polymorphisms Modify Susceptibility to Developing COVID-19. Journal of Personalized Medicine. 2022; 12(3):458. https://doi.org/10.3390/jpm12030458
Chicago/Turabian StyleDjukic, Tatjana, Goran Stevanovic, Vesna Coric, Zoran Bukumiric, Marija Pljesa-Ercegovac, Marija Matic, Djurdja Jerotic, Nevena Todorovic, Milika Asanin, Marko Ercegovac, and et al. 2022. "GSTO1, GSTO2 and ACE2 Polymorphisms Modify Susceptibility to Developing COVID-19" Journal of Personalized Medicine 12, no. 3: 458. https://doi.org/10.3390/jpm12030458
APA StyleDjukic, T., Stevanovic, G., Coric, V., Bukumiric, Z., Pljesa-Ercegovac, M., Matic, M., Jerotic, D., Todorovic, N., Asanin, M., Ercegovac, M., Ranin, J., Milosevic, I., Savic-Radojevic, A., & Simic, T. (2022). GSTO1, GSTO2 and ACE2 Polymorphisms Modify Susceptibility to Developing COVID-19. Journal of Personalized Medicine, 12(3), 458. https://doi.org/10.3390/jpm12030458