Prediction of All-Cause Mortality Based on Stress/Rest Myocardial Perfusion Imaging (MPI) Using Deep Learning: A Comparison between Image and Frequency Spectra as Input
Abstract
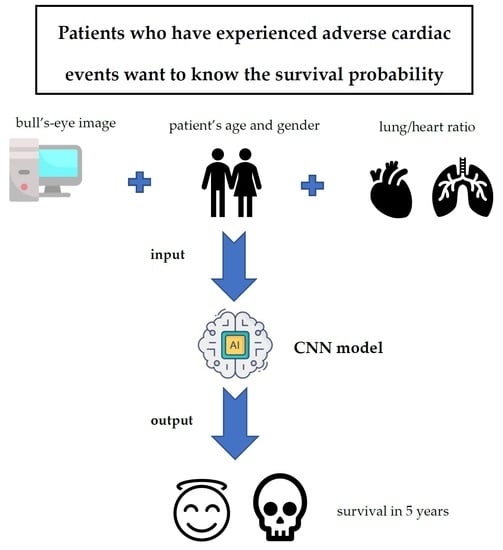
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Image Preprocessing
2.2.2. Convolutional Neural Networks (CNNs)
2.2.3. Frequency as Input
3. Results
3.1. Image Preprocessing
3.2. Results of CNNs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.who.int/news/item/09-12-2020-who-reveals-leading-causes-of-death-and-disability-worldwide-2000-2019 (accessed on 6 January 2021).
- Yannopoulos, D.; Bartos, J.A.; Aufderheide, T.P.; Callaway, C.W.; Deo, R.; Garcia, S.; Haoperin, H.R.; Kern, K.B.; Kudenchuk, P.J.; Neumar, R.W.; et al. The evolving role of the cardiac catheterization laboratory in the management of patients with out-of-hospital cardiac arrest: A scientific statement from the American heart association. Circulation 2019, 139, e530–e552. [Google Scholar] [CrossRef]
- Ahmed, T. The role of revascularization in chronic stable angina: Do we have an answer? Cureus 2020, 12, e8210. [Google Scholar] [CrossRef]
- Ora, M.; Gambhir, S. Myocardial Perfusion Imaging: A brief review of nuclear and nonnuclear techniques and comparative evaluation of recent advances. Indian J. Nucl. Med. 2019, 34, 263–270. [Google Scholar]
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J.; et al. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: A multicenter study. JACC Cardiovasc. Imaging 2018, 1, 1654–1663. [Google Scholar] [CrossRef]
- Spier, N.; Nekolla, S.; Rupprecht, C.; Mustafa, M.; Navab, N.; Baust, M. Classification of Polar Maps from Cardiac Perfusion Imaging with Graph-Convolutional Neural Networks. Sci. Rep. 2019, 9, 7569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruna, J.; Zaremba, W.; Szlam, A.; LeCun, Y. Spectral networks and locally connected networks on graphs. arXiv 2013, arXiv:1312.6203. [Google Scholar]
- Henaff, M.; Bruna, J.; LeCun, Y. Deep convolutional networks on graph-structured data. arXiv 2015, arXiv:1506.05163. [Google Scholar]
- Betancur, J.; Otaki, Y.; Motwani, M.; Fish, M.B.; Lemley, M.; Dey, D.; Gransar, H.; Tamarappoo, B.; Germano, G.; Sharir, T.; et al. Prognostic Value of Combined Clinical and Myocardial Perfusion Imaging Data Using Machine Learning. JACC: Cardiovasc. Imaging 2018, 11, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.H.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q.; et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicenter prospective registry analysis. Eur. Heart J. 2017, 38, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.A.; Payrovnaziri, S.N.; Bian, J.; He, Z. Building computational models to predict one-year mortality in ICU patients with acute myocardial infarction and post myocardial infarction syndrome. AMIA Summits Transl. Sci. Proc. 2019, 2019, 407. [Google Scholar]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.-W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.P.; Parikh, J.R.; Shenfeld, D.K.; Ivanov, V.; Marks, C.; Church, B.W.; Laramie, J.M.; Mardekian, J.; Piper, B.A.; Willke, R.J.; et al. Reverse engineering and evaluation of prediction models for progression to Type 2 diabetes: An application of machine learning using electronic health records. J. Diabetes Sci. Technol. 2016, 10, 6–18. [Google Scholar] [CrossRef] [Green Version]
- Rojas, J.C.; Carey, K.A.; Edelson, D.P.; Venable, L.R.; Howell, M.D.; Churpek, M.M. Predicting intensive care unit readmission with machine learning using electronic health record data. Ann. Am. Thorac. Soc. 2018, 15, 846–853. [Google Scholar] [CrossRef]
- Zhu, F.; Panwar, B.; Dodge, H.H.; Li, H.; Hampstead, B.M.; Albin, R.L.; Paulson, H.L.; Guan, Y. COMPASS: A computational model to predict changes in MMSE scores 24-months after initial assessment of Alzheimer’s disease. Sci. Rep. 2016, 6, 34567. [Google Scholar] [CrossRef] [Green Version]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated Lung Nodule Detection and Classification Using Deep Learning Combined with Multiple Strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Long, T.; Dai, Y.; Huang, Y. Mask R-CNN-Based Detection and Segmentation for Pulmonary Nodule 3D Visualization Diagnosis. IEEE Access 2020, 8, 44400–44409. [Google Scholar] [CrossRef]
- Cheng, D.-C.; Hsieh, T.-C.; Yen, K.-Y.; Kao, C.-H. Lesion-Based Bone Metastasis Detection in Chest Bone Scintigraphy Images of Prostate Cancer Patients Using Pre-Train, Negative Mining, and Deep Learning. Diagnostics 2021, 11, 518. [Google Scholar] [CrossRef]
- Song, H.; Jin, S.; Xiang, P.; Hu, S.; Jin, J. Prognostic value of the bone scan index in patients with metastatic castration-resistant prostate cancer: A systematic review and meta-analysis. BMC Cancer 2020, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.S.; Kim, J.Y.; Jeon, E.; Choi, W.S.; Kim, N.H.; Lee, K.Y. Evaluation of scalability and degree of fine-tuning of deep convolutional neural networks for COVID-19 screening on chest X-ray images using explainable deep-learning algorithm. J. Pers. Med. 2020, 10, 213. [Google Scholar] [CrossRef]
- Suh, Y.J.; Jung, J.; Cho, B.J. Automated breast cancer detection in digital mammograms of various densities via deep learning. J. Pers. Med. 2020, 10, 211. [Google Scholar] [CrossRef]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; de Vos, B.D.; Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef]
- González, R.C.; Woods, R.E. Digital Image Processing; Prentice Hall: Hoboken, NJ, USA, 2008. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. Computer Vision and Pattern Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Available online: https://github.com/tensorflow/tensorflow/blob/v2.4.1/tensorflow/python/keras/applications/resnet_v2.py#L31-L59 (accessed on 5 May 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identify mappings in deep residual networks. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 630–645. [Google Scholar]
- Available online: https://keras.io/api/applications/resnet/#resnet101v2-function (accessed on 5 May 2021).
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. Computer Vision and Pattern Recognition. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Available online: https://github.com/tensorflow/tensorflow/blob/v2.4.1/tensorflow/python/keras/applications/mobilenet.py#L83-L312 (accessed on 5 May 2021).
- Available online: https://github.com/tensorflow/tensorflow/blob/v2.4.1/tensorflow/python/keras/applications/mobilenet_v2.py#L95-L411 (accessed on 5 May 2021).
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. arXiv 2016, arXiv:1610.02391v4. [Google Scholar]








| Epoch | Batchsize | ResNet 50V2 | ResNet 101V2 | Mobile NetV1 | Mobile NetV2 | Xception | VGG16 | EfficientNetB0 | DenseNet169 |
|---|---|---|---|---|---|---|---|---|---|
| 40 | 16 | 0.71 | 0.71 | 0.67 | 0.61 | 0.68 | 0.62 | 0.64 | 0.70 |
| 60 | 16 | 0.71 | 0.67 | 0.70 | 0.67 | 0.63 | 0.68 | 0.72 | 0.66 |
| 60 | 32 | 0.69 | 0.68 | 0.67 | 0.68 | 0.71 | 0.64 | 0.69 | 0.71 |
| 60 | 64 | 0.64 | 0.71 | 0.72 | 0.69 | 0.70 | 0.67 | 0.76 | 0.67 |
| 80 | 16 | 0.70 | 0.71 | 0.69 | 0.69 | 0.68 | 0.72 | 0.71 | 0.71 |
| 80 | 32 | 0.70 | 0.68 | 0.64 | 0.69 | 0.68 | 0.67 | 0.68 | 0.69 |
| 80 | 64 | 0.66 | 0.69 | 0.72 | 0.63 | 0.70 | 0.70 | 0.71 | 0.71 |
| 120 | 16 | 0.70 | 0.70 | 0.69 | 0.74 | 0.66 | 0.70 | 0.64 | 0.67 |
| 120 | 32 | 0.71 | 0.72 | 0.64 | 0.72 | 0.64 | 0.68 | 0.69 | 0.69 |
| 120 | 64 | 0.61 | 0.70 | 0.74 | 0.70 | 0.63 | 0.62 | 0.68 | 0.66 |
| 160 | 16 | 0.69 | 0.66 | 0.70 | 0.68 | 0.68 | 0.69 | 0.70 | 0.66 |
| 160 | 32 | 0.68 | 0.74 | 0.68 | 0.67 | 0.67 | 0.67 | 0.68 | 0.66 |
| 160 | 64 | 0.75 | 0.69 | 0.63 | 0.70 | 0.68 | 0.70 | 0.67 | 0.68 |
| Median | 0.70 | 0.70 | 0.69 | 0.69 | 0.68 | 0.68 | 0.69 | 0.68 | |
| Epoch | Batchsize | ResNet 50V2 | ResNet 101V2 | Mobile NetV1 | Mobile NetV2 | Xception | VGG16 | EfficientNetB0 | DenseNet169 |
|---|---|---|---|---|---|---|---|---|---|
| 40 | 16 | 0.76 | 0.77 | 0.77 | 0.76 | 0.71 | 0.77 | 0.77 | 0.77 |
| 80 | 32 | 0.78 | 0.77 | 0.70 | 0.76 | 0.76 | 0.76 | 0.77 | 0.77 |
| 160 | 64 | 0.77 | 0.76 | 0.77 | 0.71 | 0.76 | 0.77 | 0.76 | 0.76 |
| Median | 0.77 | 0.77 | 0.77 | 0.76 | 0.76 | 0.77 | 0.77 | 0.77 | |
| Number | 1 | 2 | 3 | 4 | 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Death | Alive | Death | Alive | Death | Alive | Death | Alive | Death | Alive | |
| Ground truth | Death | 54 | 6 | 50 | 10 | 56 | 8 | 54 | 6 | 54 | 6 |
| Alive | 11 | 15 | 13 | 13 | 12 | 10 | 13 | 13 | 11 | 15 | |
| Accuracy | 0.80 | 0.73 | 0.77 | 0.78 | 0.80 | ||||||
| Parameter Absent | Lung/Heart Ratio | Age | Gender | |
|---|---|---|---|---|
| Number | 1 | 0.79 | 0.58 | 0.62 |
| 2 | 0.77 | 0.62 | 0.70 | |
| 3 | 0.71 | 0.60 | 0.70 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, D.-C.; Hsieh, T.-C.; Hsu, Y.-J.; Lai, Y.-C.; Yen, K.-Y.; Wang, C.C.N.; Kao, C.-H. Prediction of All-Cause Mortality Based on Stress/Rest Myocardial Perfusion Imaging (MPI) Using Deep Learning: A Comparison between Image and Frequency Spectra as Input. J. Pers. Med. 2022, 12, 1105. https://doi.org/10.3390/jpm12071105
Cheng D-C, Hsieh T-C, Hsu Y-J, Lai Y-C, Yen K-Y, Wang CCN, Kao C-H. Prediction of All-Cause Mortality Based on Stress/Rest Myocardial Perfusion Imaging (MPI) Using Deep Learning: A Comparison between Image and Frequency Spectra as Input. Journal of Personalized Medicine. 2022; 12(7):1105. https://doi.org/10.3390/jpm12071105
Chicago/Turabian StyleCheng, Da-Chuan, Te-Chun Hsieh, Yu-Ju Hsu, Yung-Chi Lai, Kuo-Yang Yen, Charles C. N. Wang, and Chia-Hung Kao. 2022. "Prediction of All-Cause Mortality Based on Stress/Rest Myocardial Perfusion Imaging (MPI) Using Deep Learning: A Comparison between Image and Frequency Spectra as Input" Journal of Personalized Medicine 12, no. 7: 1105. https://doi.org/10.3390/jpm12071105
APA StyleCheng, D. -C., Hsieh, T. -C., Hsu, Y. -J., Lai, Y. -C., Yen, K. -Y., Wang, C. C. N., & Kao, C. -H. (2022). Prediction of All-Cause Mortality Based on Stress/Rest Myocardial Perfusion Imaging (MPI) Using Deep Learning: A Comparison between Image and Frequency Spectra as Input. Journal of Personalized Medicine, 12(7), 1105. https://doi.org/10.3390/jpm12071105