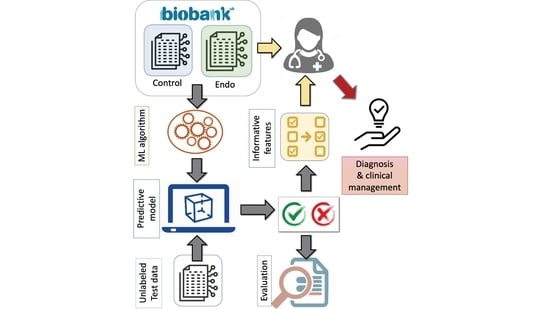
Revisiting the Risk Factors for Endometriosis: A Machine Learning Approach
Abstract
:1. Introduction
2. Methods
2.1. UKB Data Extraction and Processing
2.2. Genetic Analysis
2.3. Machine Learning Methodology
2.4. Feature Engineering
2.5. Statistical Tests
3. Results
3.1. Unification of Data from UKB: Case-Control Population-Based Groups
3.2. Univariate Statistics of Control and Endometriosis Patients from the UKB
3.3. Predictive Risk Model for Endometriosis
3.4. Informative Features and Interpretability of the Combined Model
3.5. Model’s Limitation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AUC | Area Under the ROC Curve |
| DL | Deep Learning |
| HER | Electronic Health Records |
| OT | Open Targets |
| ROC | Receiver Operating Characteristic Curve |
| IBS | Irritable bowel syndrome |
| UKB | UK-Biobank |
| PRS | Polygenic Risk Score |
| T2D | Type 2 Diabetes |
| BMI | Body Mass Index |
References
- Giudice, L.C. Clinical practice. Endometriosis. N. Engl. J. Med. 2010, 362, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Lebovic, D.I.; Mueller, M.D.; Taylor, R.N. Immunobiology of endometriosis. Fertil. Steril. 2001, 75, 1–10. [Google Scholar] [CrossRef]
- Morotti, M.; Vincent, K.; Brawn, J.; Zondervan, K.T.; Becker, C.M. Peripheral changes in endometriosis-associated pain. Hum. Reprod. Update 2014, 20, 717–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkley, K.J.; Rapkin, A.J.; Papka, R.E. The pains of endometriosis. Science 2005, 308, 1587–1589. [Google Scholar] [CrossRef]
- Meuleman, C.; Vandenabeele, B.; Fieuws, S.; Spiessens, C.; Timmerman, D.; D’Hooghe, T. High prevalence of endometriosis in infertile women with normal ovulation and normospermic partners. Fertil. Steril. 2009, 92, 68–74. [Google Scholar] [CrossRef]
- Soliman, A.M.; Fuldeore, M.; Snabes, M.C. Factors associated with time to endometriosis diagnosis in the United States. J. Women’s Health 2017, 26, 788–797. [Google Scholar] [CrossRef]
- Agarwal, S.K.; Chapron, C.; Giudice, L.C.; Laufer, M.R.; Leyland, N.; Missmer, S.A.; Singh, S.S.; Taylor, H.S. Clinical diagnosis of endometriosis: A call to action. Am. J. Obstet. Gynecol. 2019, 220, 354.e1–354.e12. [Google Scholar] [CrossRef] [Green Version]
- Denny, E.; Mann, M.C.H. A clinical overview of endometriosis: A misunderstood disease. Br. J. Nurs. 2007, 16, 1112–1116. [Google Scholar] [CrossRef]
- Brosens, I.; Benagiano, G. Endometriosis, a modern syndrome. Indian J. Med. Res. 2011, 133, 581–593. [Google Scholar]
- Ghiasi, M.; Kulkarni, M.T.; Missmer, S.A. Is Endometriosis More Common and More Severe Than It Was 30 Years Ago? J. Minim. Invasive Gynecol. 2020, 27, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, R.; Mardon, H.; Barlow, D.; Kennedy, S. Delay in the diagnosis of endometriosis: A survey of women from the USA and the UK. Hum. Reprod. 1996, 11, 878–880. [Google Scholar] [CrossRef] [Green Version]
- Husby, G.K.; Haugen, R.S.; Moen, M.H. Diagnostic delay in women with pain and endometriosis. Acta Obstet. Et. Gynecol. Scand. 2003, 82, 649–653. [Google Scholar] [CrossRef]
- Ballard, K.; Lowton, K.; Wright, J. What’s the delay? A qualitative study of women’s experiences of reaching a diagnosis of endometriosis. Fertil. Steril. 2006, 86, 1296–1301. [Google Scholar] [CrossRef]
- Nnoaham, K.E.; Hummelshoj, L.; Webster, P.; d’Hooghe, T.; de Cicco Nardone, F.; de Cicco Nardone, C.; Jenkinson, C.; Kennedy, S.H.; Zondervan, K.T.; World Endometriosis Research Foundation Global Study of Women’s Health. Impact of endometriosis on quality of life and work productivity: A multicenter study across ten countries. Fertil. Steril. 2011, 96, 366–373.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scioscia, M.; Virgilio, B.A.; Laganà, A.S.; Bernardini, T.; Fattizzi, N.; Neri, M.; Guerriero, S. Differential diagnosis of endometriosis by ultrasound: A rising challenge. Diagnostics 2020, 10, 848. [Google Scholar] [CrossRef] [PubMed]
- Kiesel, L.; Sourouni, M. Diagnosis of endometriosis in the 21st century. Climacteric 2019, 22, 296–302. [Google Scholar] [CrossRef]
- Zakhari, A.; Delpero, E.; McKeown, S.; Tomlinson, G.; Bougie, O.; Murji, A. Endometriosis recurrence following post-operative hormonal suppression: A systematic review and meta-analysis. Hum. Reprod. Update 2021, 27, 96–107. [Google Scholar] [CrossRef]
- Schwartz, K.; Llarena, N.C.; Rehmer, J.M.; Richards, E.G.; Falcone, T. The role of pharmacotherapy in the treatment of endometriosis across the lifespan. Expert Opin. Pharm. 2020, 21, 893–903. [Google Scholar] [CrossRef]
- Kalaitzopoulos, D.R.; Samartzis, N.; Kolovos, G.N.; Mareti, E.; Samartzis, E.P.; Eberhard, M.; Dinas, K.; Daniilidis, A. Treatment of endometriosis: A review with comparison of 8 guidelines. BMC Womens Health 2021, 21, 397. [Google Scholar] [CrossRef] [PubMed]
- Marinho, M.C.; Magalhaes, T.F.; Fernandes, L.F.C.; Augusto, K.L.; Brilhante, A.V.; Bezerra, L.R. Quality of life in women with endometriosis: An integrative review. J. Women’s Health 2018, 27, 399–408. [Google Scholar] [CrossRef]
- Parasar, P.; Ozcan, P.; Terry, K.L. Endometriosis: Epidemiology, diagnosis and clinical management. Curr. Obstet. Gynecol. Rep. 2017, 6, 34–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, D.K.; Correia, K.F.; Vitonis, A.F.; Missmer, S.A. Body size and endometriosis: Results from 20 years of follow-up within the Nurses’ Health Study II prospective cohort. Hum. Reprod. 2013, 28, 1783–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, D.W.; Missmer, S.A. The epidemiology of endometriosis. Ann. N. Y. Acad. Sci. 2002, 955, 11–22. [Google Scholar] [CrossRef]
- Missmer, S.A.; Chavarro, J.E.; Malspeis, S.; Bertone-Johnson, E.R.; Hornstein, M.D.; Spiegelman, D.; Barbieri, R.L.; Willett, W.C.; Hankinson, S.E. A prospective study of dietary fat consumption and endometriosis risk. Hum. Reprod. 2010, 25, 1528–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, C.M.; Johnstone, E.B.; Hammoud, A.O.; Stanford, J.B.; Varner, M.W.; Kennedy, A.; Chen, Z.; Sun, L.; Fujimoto, V.Y.; Hediger, M.L.; et al. Risk factors associated with endometriosis: Importance of study population for characterizing disease in the ENDO Study. Am. J. Obs. Gynecol. 2013, 208, 451.e1–451.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghese, B.; Zondervan, K.T.; Abrao, M.S.; Chapron, C.; Vaiman, D. Recent insights on the genetics and epigenetics of endometriosis. Clin. Genet. 2017, 91, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augoulea, A.; Alexandrou, A.; Creatsa, M.; Vrachnis, N.; Lambrinoudaki, I. Pathogenesis of endometriosis: The role of genetics, inflammation and oxidative stress. Arch. Gynecol. Obstet. 2012, 286, 99–103. [Google Scholar] [CrossRef]
- Sapkota, Y.; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; Jones, S.; et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef] [Green Version]
- Fung, J.N.; Rogers, P.A.; Montgomery, G.W. Identifying the biological basis of GWAS hits for endometriosis. Biol. Reprod. 2015, 92, 87. [Google Scholar] [CrossRef] [Green Version]
- Albertsen, H.M.; Ward, K. Genes linked to endometriosis by GWAS are integral to cytoskeleton regulation and suggests that mesothelial barrier homeostasis is a factor in the pathogenesis of endometriosis. Reprod. Sci. 2017, 24, 803–811. [Google Scholar] [CrossRef]
- Ahn, S.H.; Khalaj, K.; Young, S.L.; Lessey, B.A.; Koti, M.; Tayade, C. Immune-inflammation gene signatures in endometriosis patients. Fertil. Steril. 2016, 106, 1420–1431.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, P.T.K.; Horne, A.W. Endometriosis: Etiology, pathobiology, and therapeutic prospects. Cell 2021, 184, 2807–2824. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canela-Xandri, O.; Rawlik, K.; Tenesa, A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018, 50, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Silva, D.; Pierleoni, A.; Pignatelli, M.; Ong, C.; Fumis, L.; Karamanis, N.; Carmona, M.; Faulconbridge, A.; Hercules, A.; McAuley, E. Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 2019, 47, D1056–D1065. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prive, F.; Aschard, H.; Carmi, S.; Folkersen, L.; Hoggart, C.; O’Reilly, P.F.; Vilhjalmsson, B.J. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet. 2022, 109, 12–23. [Google Scholar] [CrossRef]
- Dorogush, A.V.; Ershov, V.; Gulin, A. CatBoost: Gradient boosting with categorical features support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Hancock, J.T.; Khoshgoftaar, T.M. CatBoost for big data: An interdisciplinary review. J. Big Data 2020, 7, 1–45. [Google Scholar] [CrossRef]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. Adv. Neural Inf. Processing Syst. 2018, 31. [Google Scholar]
- Vercellini, P.; Fedele, L.; Aimi, G.; Pietropaolo, G.; Consonni, D.; Crosignani, P. Association between endometriosis stage, lesion type, patient characteristics and severity of pelvic pain symptoms: A multivariate analysis of over 1000 patients. Hum. Reprod. 2007, 22, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Tanbo, T.; Fedorcsak, P. Endometriosis-associated infertility: Aspects of pathophysiological mechanisms and treatment options. Acta Obstet. Gynecol. Scand. 2017, 96, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.S.; DiVasta, A.D.; Vitonis, A.F.; Sarda, V.; Laufer, M.R.; Missmer, S.A. The impact of endometriosis on quality of life in adolescents. J. Adolesc. Health 2018, 63, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Grogan, S.; Turley, E.; Cole, J. ‘So many women suffer in silence’: A thematic analysis of women’s written accounts of coping with endometriosis. Psychol. Health 2018, 33, 1364–1378. [Google Scholar] [CrossRef] [PubMed]
- Králíčková, M.; Laganà, A.S.; Ghezzi, F.; Vetvicka, V. Endometriosis and risk of ovarian cancer: What do we know? Arch. Gynecol. Obstet. 2020, 301, 1–10. [Google Scholar] [CrossRef]
- Heidemann, L.N.; Hartwell, D.; Heidemann, C.H.; Jochumsen, K.M. The relation between endometriosis and ovarian cancer—A review. Acta Obs. Gynecol. Scand. 2014, 93, 20–31. [Google Scholar] [CrossRef]
- Anastasiu, C.V.; Moga, M.A.; Elena Neculau, A.; Bălan, A.; Scârneciu, I.; Dragomir, R.M.; Dull, A.-M.; Chicea, L.-M. Biomarkers for the noninvasive diagnosis of endometriosis: State of the art and future perspectives. Int. J. Mol. Sci. 2020, 21, 1750. [Google Scholar] [CrossRef] [Green Version]
- Fassbender, A.; Burney, R.O.; O, D.F.; D’Hooghe, T.; Giudice, L. Update on Biomarkers for the Detection of Endometriosis. Biomed. Res. Int. 2015, 2015, 130854. [Google Scholar] [CrossRef] [Green Version]
- Chapron, C.; Lafay-Pillet, M.C.; Santulli, P.; Bourdon, M.; Maignien, C.; Gaudet-Chardonnet, A.; Maitrot-Mantelet, L.; Borghese, B.; Marcellin, L. A new validated screening method for endometriosis diagnosis based on patient questionnaires. EClinicalMedicine 2022, 44, 101263. [Google Scholar] [CrossRef]
- Streuli, I.; Gaitzsch, H.; Wenger, J.M.; Petignat, P. Endometriosis after menopause: Physiopathology and management of an uncommon condition. Climacteric 2017, 20, 138–143. [Google Scholar] [CrossRef]
- Akter, S.; Xu, D.; Nagel, S.C.; Bromfield, J.J.; Pelch, K.E.; Wilshire, G.B.; Joshi, T. GenomeForest: An Ensemble Machine Learning Classifier for Endometriosis. AMIA Jt. Summits. Transl. Sci. Proc. 2020, 2020, 33–42. [Google Scholar] [PubMed]
- Viganò, D.; Zara, F.; Usai, P. Irritable bowel syndrome and endometriosis: New insights for old diseases. Dig. Liver Dis. 2018, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Chiaffarino, F.; Cipriani, S.; Ricci, E.; Mauri, P.A.; Esposito, G.; Barretta, M.; Vercellini, P.; Parazzini, F. Endometriosis and irritable bowel syndrome: A systematic review and meta-analysis. Arch. Gynecol. Obs. 2021, 303, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Nilufer, R.; Karina, B.; Paraskevi, C.; Rebecca, D.; Genevieve, G.; Ayush, G.; Stuart, M.; Sally, M.; Yadav, S.; Andrew, S.J. Large-scale genome-wide association meta-analysis of endometriosis reveals 13 novel loci and genetically-associated comorbidity with other pain conditions. BioRxiv 2018, 406967. [Google Scholar] [CrossRef] [Green Version]
- Bischoff, F.; Simpson, J.L. Genetics of endometriosis: Heritability and candidate genes. Best Pract. Res. Clin. Obstet. Gynaecol. 2004, 18, 219–232. [Google Scholar] [CrossRef]
- Nyholt, D.R.; Low, S.K.; Anderson, C.A.; Painter, J.N.; Uno, S.; Morris, A.P.; MacGregor, S.; Gordon, S.D.; Henders, A.K.; Martin, N.G.; et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat. Genet. 2012, 44, 1355–1359. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Sapkota, Y.; Fung, J.; Montgomery, G.W. Genetic biomarkers for endometriosis. In Biomarkers for Endometriosis; Springer: Berlin/Heidelberg, Germany, 2017; pp. 83–93. [Google Scholar]
- Kloeve-Mogensen, K.; Rohde, P.D.; Twisttmann, S.; Nygaard, M.; Koldby, K.M.; Steffensen, R.; Dahl, C.M.; Rytter, D.; Overgaard, M.T.; Forman, A. Polygenic Risk Score Prediction for Endometriosis. Front. Reprod. Health 2021, 3, 793226. [Google Scholar] [CrossRef]
- Moldovan, A.; Waldman, Y.Y.; Brandes, N.; Linial, M. Body Mass Index and Birth Weight Improve Polygenic Risk Score for Type 2 Diabetes. J. Pers. Med. 2021, 11, 582. [Google Scholar] [CrossRef]
- Mowers, E.L.; Lim, C.S.; Skinner, B.; Mahnert, N.; Kamdar, N.; Morgan, D.M.; As-Sanie, S. Prevalence of endometriosis during abdominal or laparoscopic hysterectomy for chronic pelvic pain. Obstet. Gynecol. 2016, 127, 1045–1053. [Google Scholar] [CrossRef]
- Visalaxi, S.; Punnoose, D.; Muthu, T.S. An analogy of endometriosis recognition using machine learning techniques. In Proceedings of the 2021 Third International Conference on Intelligent Communication Technologies and Virtual Mobile Networks (ICICV), Tirunelveli, India, 4–6 February 2021; pp. 739–746. [Google Scholar]
- Bendifallah, S.; Puchar, A.; Suisse, S.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Touboul, C.; Dabi, Y.; Daraï, E. Machine learning algorithms as new screening approach for patients with endometriosis. Sci. Rep. 2022, 12, 639. [Google Scholar] [CrossRef]







| Attributes & Traits (Units) | Data Type Class | UKB Field ID | Number of Women | Missing Data (%) | Mean [Cardinality] |
|---|---|---|---|---|---|
| Body mass index (BMI) | Physical measures | 21001 | 148,026 | <1 | 27.2 |
| Smoking | Lifestyle & environment | 20116 | 37,444 | 74.8 | [4] |
| Birth weight (kg) | Early life factors | 20022 | 52,645 | 35.5 | 3.32 |
| Number of live birth | Female-specific factors | 2734 | 148,402 | <1 | 1.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blass, I.; Sahar, T.; Shraibman, A.; Ofer, D.; Rappoport, N.; Linial, M. Revisiting the Risk Factors for Endometriosis: A Machine Learning Approach. J. Pers. Med. 2022, 12, 1114. https://doi.org/10.3390/jpm12071114
Blass I, Sahar T, Shraibman A, Ofer D, Rappoport N, Linial M. Revisiting the Risk Factors for Endometriosis: A Machine Learning Approach. Journal of Personalized Medicine. 2022; 12(7):1114. https://doi.org/10.3390/jpm12071114
Chicago/Turabian StyleBlass, Ido, Tali Sahar, Adi Shraibman, Dan Ofer, Nadav Rappoport, and Michal Linial. 2022. "Revisiting the Risk Factors for Endometriosis: A Machine Learning Approach" Journal of Personalized Medicine 12, no. 7: 1114. https://doi.org/10.3390/jpm12071114
APA StyleBlass, I., Sahar, T., Shraibman, A., Ofer, D., Rappoport, N., & Linial, M. (2022). Revisiting the Risk Factors for Endometriosis: A Machine Learning Approach. Journal of Personalized Medicine, 12(7), 1114. https://doi.org/10.3390/jpm12071114