Detection of Paroxysmal Atrial Fibrillation from Dynamic ECG Recordings Based on a Deep Learning Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Databases
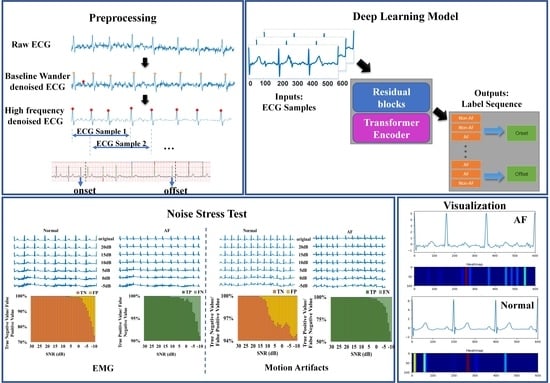
2.2. Data Preprocessing
2.3. Methods
3. Results
3.1. Model Training
3.2. Metrics
3.3. Model Evaluation and Ablation Experiments
3.4. Performance in AF Rhythm Discrimination and Classification
3.5. Performance in the Detection of AF Onsets, Offsets, and Episodes
3.6. Performance on CPSC2021 Hidden Test Data
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michael, E. Field. Chapter 35-Atrial Fibrillation. In Cardiology Secrets, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 323–329. [Google Scholar]
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics-2019 Update: A Report from the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef]
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.-H.; McAnulty, J.H., Jr.; Zheng, Z.-J.; et al. Worldwide epidemiology of atrial fibrillation: A Global Burden of Disease 2010 Study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Colilla, S.; Crow, A.; Petkun, W.; Singer, D.E.; Simon, T.; Liu, X. Estimates of current and future incidence and prevalence of atrial fibrillation in the U.S. adult population. Am. J. Cardiol. 2013, 112, 1142–1147. [Google Scholar] [CrossRef]
- Krijthe, B.P.; Kunst, A.; Benjamin, E.J.; Lip, G.Y.; Franco, O.H.; Hofman, A.; Witteman, J.C.; Stricker, B.H.; Heeringa, J. Projections on the number of individuals with atrial fibrillation in the European Union, from 2000 to 2060. Eur. Heart J. 2013, 34, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.S.; O’Connell, H.; Li, S.; Ziegler, P.D. Thirty-Second Gold Standard Definition of Atrial Fibrillation and Its Relationship with Subsequent Arrhythmia Patterns: Analysis of a Large Prospective Device Database. Circ. Arrhythm. Electrophysiol. 2018, 11, e006274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, C.; Wei, S.; Shen, Q.; Zhou, F.; Li, J. A New Entropy-Based Atrial Fibrillation Detection Method for Scanning Wearable ECG Recordings. Entropy 2018, 20, 904. [Google Scholar] [CrossRef] [PubMed]
- Kalidas, V.; Tamil, L.S. Detection of atrial fibrillation using discrete-state Markov models and Random Forests. Comput. Biol. Med. 2019, 113, 103386. [Google Scholar] [CrossRef]
- Parsi, A.; Glavin, M.; Jones, E.; Byrne, D. Prediction of paroxysmal atrial fibrillation using new heart rate variability features. Comput. Biol. Med. 2021, 133, 104367. [Google Scholar] [CrossRef]
- Pürerfellner, H.; Pokushalov, E.; Sarkar, S.; Koehler, J.; Zhou, R.; Urban, L.; Hindricks, G. P-wave evidence as a method for improving algorithm to detect atrial fibrillation in insertable cardiac monitors. Heart Rhythm. 2014, 11, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Ladavich, S.; Ghoraani, B. Rate-independent detection of atrial fibrillation by statistical modeling of atrial activity. Biomed. Signal Process. Control. 2018, 18, 274–281. [Google Scholar] [CrossRef]
- Mukherjee, A.; Choudhury, A.D.; Datta, S.; Puri, C.; Banerjee, R.; Singh, R.; Ukil, A.; Bandyopadhyay, S.; Pal, A.; Khandelwal, S. Detection of atrial fibrillation and other abnormal rhythms from ECG using a multi-layer classifier architecture. Physiol. Meas. 2019, 40, 054006. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wulan, N.; Wang, K.; Zhang, H. Detecting atrial fibrillation by deep convolutional neural networks. Comput. Biol. Med. 2018, 93, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.; Afghah, F.; Acharya, U.R. HAN-ECG: An interpretable atrial fibrillation detection model using hierarchical attention networks. Comput. Biol. Med. 2020, 127, 104057. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Shenfield, A.; Kareem, M.; San, T.R.; Fujita, H.; Acharya, U.R. Automated detection of atrial fibrillation using long short-term memory network with RR interval signals. Comput. Biol. Med. 2018, 102, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Chen, Y.; Guo, J.; Han, B.; Shi, Y.; Ji, L.; Wang, J.; Zhang, G.; Luo, J. Accurate detection of atrial fibrillation from 12-lead ECG using deep neural network. Comput. Biol. Med. 2020, 116, 103378. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Hu, Z.; Wang, R.; Yin, L.; Li, Y.; Cai, Y. A novel hybrid network of fusing rhythmic and morphological features for atrial fibrillation detection on mobile ECG signals. Neural Comput. Applic. 2020, 32, 8101–8113. [Google Scholar] [CrossRef]
- Baalman, S.W.E.; Schroevers, F.E.; Oakley, A.J.; Brouwer, T.F.; van der Stuijt, W.; Bleijendaal, H.; Ramos, L.A.; Lopes, R.R.; Marquering, H.A.; Knops, R.E.; et al. A morphology based deep learning model for atrial fibrillation detection using single cycle electrocardiographic samples. Int. J. Cardiol. 2020, 316, 130–136. [Google Scholar] [CrossRef]
- Wang, X.; Ma, C.; Zhang, X.; Gao, H.; Clifford, G.D.; Liu, C. Paroxysmal Atrial Fibrillation Events Detection from Dynamic ECG Recordings: The 4th China Physiological Signal Challenge 2021. PhysioNet. 2021. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Petrutiu, S.; Sahakian, A.V.; Swiryn, S. Abrupt changes in fibrillatory wave characteristics at the termination of paroxysmal atrial fibrillation in humans. Europace 2007, 9, 466–470. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. A new method for detecting atrial fibrillation using R-R intervals. Comput. Cardiol. 1983, 10, 227–230. [Google Scholar]
- Clifford, G.D.; Liu, C.; Moody, B.; Li-wei, H.L.; Silva, I.; Li, Q.; Johnson, A.E.; Mark, R.G. AF classification from a short single lead ECG recording: The PhysioNet/computing in cardiology challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA; pp. 6000–6010. [Google Scholar]
- Ott, M.; Edunov, S.; Grangier, D.; Auli, M. Scaling neural machine translation. In Proceedings of the Third Conference on Machine Translation: Research Papers, Brussels, Belgium, October 2018; Workshop on Statistical Machine Translation (WMT). Available online: https://aclanthology.org/W18-6301 (accessed on 9 May 2023).
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jegou, H. Training data-efficient image transformers & distillation through attention. arXiv 2020, arXiv:2012.12877. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. arXiv 2020, arXiv:2005.12872. [Google Scholar]
- Zhu, X.; Su, W.; Lu, L.; Li, B.; Wang, X.; Dai, J. Deformable DETR: Deformable transformers for end-to-end object detection. arXiv 2020, arXiv:2010.04159. [Google Scholar]
- Ye, L.; Rochan, M.; Liu, Z.; Wang, Y. Cross-modal self-attention network for referring image segmentation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Yan, G.; Liang, S.; Zhang, Y.; Liu, F. Fusing Transformer Model with Temporal Features for ECG Heartbeat Classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 898–905. [Google Scholar]
- Meng, L.; Tan, W.; Ma, J.; Wang, R.; Yin, X.; Zhang, Y. Enhancing dynamic ECG heartbeat classification with lightweight transformer model. Artif. Intell. Med. 2022, 124, 102236. [Google Scholar] [CrossRef]
- Hu, R.; Chen, J.; Zhou, L. A transformer-based deep neural network for arrhythmia detection using continuous ECG signals. Comput. Biol. Med. 2022, 144, 105325. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Prabhakararao, E.; Dandapat, S. Atrial Fibrillation Burden Estimation Using Multi-Task Deep Convolutional Neural Network. IEEE J. Biomed. Health Inform. 2022, 26, 5992–6002. [Google Scholar] [CrossRef]
- Chocron, A.; Oster, J.; Biton, S.; Mandel, F.; Elbaz, M.; Zeevi, Y.Y.; Behar, J.A. Remote atrial fibrillation burden estimation using deep recurrent neural network. IEEE Trans. Biomed. Eng. 2021, 68, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.S.; Peimankar, A.; Puthusserypady, S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst. Appl. 2019, 115, 465–473. [Google Scholar] [CrossRef]
- Tuboly, G.; Kozmann, G.; Kiss, O.; Merkely, B. Atrial fibrillation detection with and without atrial activity analysis using lead-I mobile ECG technology. Biomed. Signal Process. Control 2021, 66, 102462. [Google Scholar] [CrossRef]
- Kumar, D.; Peimankar, A.; Sharma, K.; Domínguez, H.; Puthusserypady, S.; Bardram, J.E. Deepaware: A hybrid deep learning and context-aware heuristics-based model for atrial fibrillation detection. Comput. Methods Programs Biomed. 2022, 221, 106899. [Google Scholar] [CrossRef]
- Moody, G.B.; Muldrow, W.E.; Mark, R.G. A noise stress test for arrhythmia detectors. Comput. Cardiol. 1984, 11, 381–384. [Google Scholar]
- Butkuviene, M.; Petrenas, A.; Solosenko, A.; Martin-Yebra, A.; Marozas, V.; Sornmo, L. Considerations on Performance Evaluation of Atrial Fibrillation Detectors. IEEE Trans. Bio-Med. Eng. 2021, 68, 3250–3260. [Google Scholar] [CrossRef]
- Salinas-Martínez, R.; de Bie, J.; Marzocchi, N.; Sandberg, F. Detection of Brief Episodes of Atrial Fibrillation Based on Electrocardiomatrix and Convolutional Neural Network. Front. Physiol. 2021, 12, 673–819. [Google Scholar] [CrossRef]
- Jin, Y.; Qin, C.; Huang, Y.; Zhao, W.; Liu, C. Multi-domain modeling of atrial fibrillation detection with twin attentional convolutional long short-term memory neural networks. Know.-Based Syst. 2020, 193, 105460. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated Atrial Fibrillation Detection using a Hybrid CNN-LSTM Network on Imbalanced ECG Datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]















| CPSC 2021 | AFDB | LTAF | MITDB | CinC 2017 | |
|---|---|---|---|---|---|
| Non-AF Records | 732 | 0 | 1 | 40 | 7757 |
| Persistent AF Records | 475 | 2 | 8 | 0 | / |
| Paroxysmal AF Records | 229 | 21 | 73 | 8 | / |
| Average Duration of Record | 0.3 h | 10 h | 24~25 h | 0.5 h | / |
| Average Beats in a Record | 1356 | 49,068 | 107,810 | 2346 | / |
| Duration of Record | 8 s~6 h | 10 h | 24~25 h | 0.5 h | 9~60 s |
| Total Beats | 2,146,915 | 1,128,561 | 9,055,636 | 112,646 | / |
| Subjects | 49 AF (23 PAF) 56 non-AF | / | / | 47 | / |
| Episodes with Paroxysmal AF | 677 (≥5 beats) | 285 | 7358 | 122 | / |
| Beats with AF | 770,396 | 520,394 | 3,118,292 | 13,259 | / |
| Number of Records | 1436 | 23 (2 unavailable) | 84 | 48 | 8528 |
| Sample Rate (Hz) | 200 | 250 | 128 | 360 | 300 |
| Label (Types of Rhythm) | AF/AFL/normal | AFIB/AFL/ J/Others | 9 types | 15 types | Normal/ AF/ Others/ noise |
| Lead | I, II | / | / | MLII, V1, V2, V4, V5 | / |
| Sources | 12-lead Holter or 3-lead wearable ECG monitoring devices | ambulatory ECG recorder | / | 24 h ambulatory ECG | AliveCor device |
| Layer | Output Shape | Parameters |
|---|---|---|
| Inputs | (600, 1) | 0 |
| Conv1D | (600, 32) | 544 |
| Batch Normalization | (600, 32) | 128 |
| ReLU | (600, 32) | 0 |
| Conv1D in Block 1–Block 4 | (none, 32) | 16,416 |
| Batch Normalization in Block 1–Block 4 | (none, 32) | 128 |
| Conv1D in Block 5–Block 8 | (none, 64) | 65,600 |
| Batch Normalization in Block 5–Block 8 | (none, 64) | 256 |
| Position Encoding | (38, 64) | 3200 |
| Transformer Encoder | (38, 64) | 21,088 |
| Global Average Pooling1D | (none, 64) | 0 |
| Dense | (none, 32) | 2080 |
| Dropout | (none, 32) | 0 |
| Dense | (none, 1) | 33 |
| Outputs | (none, 1) | 0 |
| Total Parameters | 653,505 | |
| Trainable Parameters | 651,905 |
| Non-AF | AF | Total of Samples | |
|---|---|---|---|
| Normal | 1,258,848 | 0 | 1,258,848 |
| Persistent AF | 0 | 675,477 | 675,477 |
| Paroxysmal AF | 117,442 | 94,919 | 212,590 |
| Total Samples | 1,375,558 | 769,921 | 2,145,479 |
| Total (After Augmentation) | 1,375,558 | 1,445,873 | 2,821,431 |
| Prediction | ||
|---|---|---|
| Real Label | Positive | Negative |
| Positive | TP (True Positive) | FN (False Negative) |
| Negative | FP (False Positive) | TN (True Negative) |
| Model | Accuracy (%) | Sensitivity (%) | Specificity (%) | FPR (%) | F1- Score | Params | Training Consumption | Testing Consumption |
|---|---|---|---|---|---|---|---|---|
| 8 Res Blocks | 97.83 | 98.80 | 97.08 | 2.92 | 0.9753 | 627,617 | 31.3 h | 45 min |
| 4 Res Blocks + Transformer | 97.98 | 98.84 | 97.32 | 2.68 | 0.9771 | 323,073 | 29.8 h | 40 min |
| 8 Res Blocks + Transformer | 98.15 | 98.06 | 98.31 | 1.59 | 0.9851 | 651,905 | 33.6 h | 45 min |
| Database | Records | Accuracy (%) | Sensitivity (%) | Specificity (%) | FPR (%) | F1-Score | |
|---|---|---|---|---|---|---|---|
| CPSC2021 | 189 | 98.15 | 98.06 | 98.31 | 1.59 | 0.9851 | |
| MITDB | CH1 | 48 | 98.32 | 80.57 | 95.32 | 4.67 | 0.8539 |
| CH2 | 48 | 95.71 | 65.07 | 97.00 | 3.00 | 0.7073 | |
| LTAF | CH1 | 84 | 82.30 | 70.35 | 84.82 | 15.18 | 0.7401 |
| CH2 | 84 | 76.67 | 64.88 | 71.33 | 28.66 | 0.6292 | |
| AFDB | CH1 | 23 | 97.91 | 90.05 | 97.03 | 2.97 | 0.8828 |
| CH2 | 23 | 98.65 | 90.12 | 97.78 | 2.22 | 0.8999 | |
| CinC2017 | 8528 | 89.29 | 79.84 | 91.09 | 8.81 | 0.7048 |
| Database | Records | Accuracy (%) | Sensitivity (%) | Specificity (%) | FPR (%) | F1-Score |
|---|---|---|---|---|---|---|
| MITDB | 48 | 96.34 | 80.57 | 97.99 | 2.00 | 0.8539 |
| LTAF | 84 | 86.16 | 65.71 | 83.09 | 16.91 | 0.7401 |
| AFDB | 23 | 98.67 | 87.69 | 98.56 | 1.44 | 0.9008 |
| Database | MITDB | AFDB | LTAF |
|---|---|---|---|
| Episodes | 122 | 285 | 7358 |
| Detected Onset | 117 | 260 | 4709 |
| Detected Offset | 107 | 227 | 4424 |
| SeOnset (%) | 95.90 | 91.23 | 64.00 |
| SeOffset (%) | 87.70 | 79.65 | 60.13 |
| Database | AFDB | MITDB | LTAF | |||
|---|---|---|---|---|---|---|
| CH1 | CH2 | CH1 | CH2 | CH1 | CH2 | |
| Episodes | 122 | 122 | 285 | 285 | 7358 | 7358 |
| Accepisode (%) | 93.34 | 93.93 | 97.69 | 96.62 | 77.41 | 68.02 |
| Seepisode (%) | 83.42 | 83.12 | 80.00 | 65.00 | 69.07 | 54.87 |
| FPRepisode(%) | 1.55 | 0.46 | 4.17 | 3.47 | 18.12 | 13.43 |
| Mccepisode | 86.35 | 76.04 | 89.75 | 63.47 | 57.97 | 58.35 |
| ECG Length | Database | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1-Score | Training Consumption | Testing Consumption (Every Sample) | |
|---|---|---|---|---|---|---|---|---|
| [16] | 10 s | CPSC 2018 | 99.35 | 99.44 | 99.19 | 0.9906 | 19.7 min | 2.7 ms |
| [43] | 10 beats | AFDB | 87.88 | 84.56 | 90.84 | 0.8686 | / | / |
| [14] | 5 s | AFDB | 98.81 | 99.08 | 98.54 | / | / | / |
| [44] | 5 s | AFDB | 98.51 | 98.14 | 98.76 | / | 122 s/epoch | 0.6 ms |
| [13] | 5 s | AFDB | 98.29 | 98.34 | 97.87 | / | 40 min | / |
| [45] | 1 beat | AFDB | / | 96.68 | 98.4 | 0.9705 | 44 s/epoch | / |
| Our method | 3 beats | AFDB | 98.69 | 87.69 | 98.56 | 0.9008 | / | 1.1 ms |
| AFDB (CH1) | 97.91 | 90.05 | 97.03 | 0.8828 | / | 0.52 ms | ||
| AFDB (CH2) | 98.65 | 90.12 | 97.78 | 0.8999 | / | 0.52 ms |
| Database | Metrics | Our Method | Salinas-Martínez et al. [24] |
|---|---|---|---|
| AFDB | SeDur (%) | 90.05–90.12 | 75.95–86.71 |
| PPVdur (%) | 85.21–88.43 | 89.85–93.40 | |
| Seepisode (%) | 83.18–83.42 | 96.73–97.45 | |
| PPVepisode (%) | 94.13–95.23 | 61.10–80.15 | |
| FPRepisode (%) | 0.46–1.55 | - | |
| Seonset (%) | 91.23 | - | |
| Seoffset (%) | 79.65 | - | |
| MITDB | Sedur (%) | 65.07–80.57 | 85.26–95.32 |
| PPVdur (%) | 26.89–29.46 | 31.05–31.50 | |
| Seepisode (%) | 65.00–80.00 | 90.65–98.13 | |
| PPVepisode (%) | 30.80–57.86 | 8.32–12.99 | |
| FPRepisode (%) | 3.47–4.17 | - | |
| Seonset (%) | 95.90 | - | |
| Seoffset (%) | 87.70 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Feng, T.; Wang, M.; Liu, C.; Tang, H. Detection of Paroxysmal Atrial Fibrillation from Dynamic ECG Recordings Based on a Deep Learning Model. J. Pers. Med. 2023, 13, 820. https://doi.org/10.3390/jpm13050820
Hu Y, Feng T, Wang M, Liu C, Tang H. Detection of Paroxysmal Atrial Fibrillation from Dynamic ECG Recordings Based on a Deep Learning Model. Journal of Personalized Medicine. 2023; 13(5):820. https://doi.org/10.3390/jpm13050820
Chicago/Turabian StyleHu, Yating, Tengfei Feng, Miao Wang, Chengyu Liu, and Hong Tang. 2023. "Detection of Paroxysmal Atrial Fibrillation from Dynamic ECG Recordings Based on a Deep Learning Model" Journal of Personalized Medicine 13, no. 5: 820. https://doi.org/10.3390/jpm13050820
APA StyleHu, Y., Feng, T., Wang, M., Liu, C., & Tang, H. (2023). Detection of Paroxysmal Atrial Fibrillation from Dynamic ECG Recordings Based on a Deep Learning Model. Journal of Personalized Medicine, 13(5), 820. https://doi.org/10.3390/jpm13050820