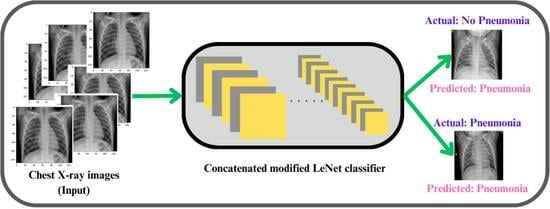
Concatenated Modified LeNet Approach for Classifying Pneumonia Images
Abstract
:1. Introduction
1.1. Pneumonia Imaging Modalities
- Variability in Image Quality: Chest X-ray image quality can vary significantly, making it challenging to identify subtle abnormalities.
- Overlapping Features: Pneumonia patterns may overlap with other lung conditions, leading to misinterpretations.
- Subjectivity: Interpretation of X-rays is subjective and relies on radiologist expertise.
- Size and Location of Infection: The size and location of pneumonia can affect its visibility in X-rays.
- Co-occurring Conditions: Patients with pneumonia may have co-occurring conditions that complicate interpretation.
- Children and Elderly Patients: Detecting pneumonia in children and the elderly can be challenging due to anatomical and age-related differences.
- Evolution of Infections: Pneumonia can evolve rapidly, and X-ray findings may change.
- Atypical Presentations: Pneumonia may present atypically, deviating from typical radiographic patterns.
- Data Imbalance: Imbalanced datasets can lead to biases in model performance.
1.2. Artificial Intelligence-Based Models for Disease Diagnosis
2. Literature Survey
2.1. Exploring Deep Learning Applications in Medical Imaging
2.2. Pneumonia Diagnosis Using Deep Learning
- Non-COVID-19 viral pneumonia vs. normal: The model demonstrated 94.43% accuracy, 98.19% sensitivity, and 95.78% specificity.
- Bacterial pneumonia vs. normal: The model achieved 91.43% accuracy, 91.94% sensitivity, and 100% specificity.
- COVID-19 vs. normal: Here, the model’s accuracy reached 99.16%, sensitivity 97.44%, and specificity 100%.
- COVID-19 vs. non-COVID-19 viral pneumonia: Accuracy was 99.62%, sensitivity 90.63%, and specificity 99.89%.
- Three-way classification: The model displayed 94.00% accuracy, 91.30% sensitivity, and 84.78% specificity.
- Four-way classification: Accuracy was 93.42%, sensitivity 89.18%, and specificity 98.92%.
2.3. Concatenated Classifiers in Medical Imaging
2.4. Challenges and Opportunities
3. Proposed Concatenated Modified LeNet-5 Model
- Input Layer: At the bottom of the diagram, a layer representing the input data with the shape (128, 128, 1).
- LeNet-5 Model (Repeated Three Times): For each Concatenated model, the following blocks, repeated three times (because nets = 3 in the proposed model):
- ➢
- Convolutional Layer: 6 filters, kernel size (5, 5), ReLU activation, with Average Pooling (pool size: 2 × 2).
- ➢
- Convolutional Layer: 16 filters, kernel size (5, 5), ReLU activation, with Average Pooling (pool size: 2 × 2).
- ➢
- Flatten Layer: Flattens the output.
- ➢
- Fully Connected Layer: 120 neurons, ReLU activation.
- ➢
- Fully Connected Layer: 84 neurons, ReLU activation.
- ➢
- Output Layer: 2 neurons with softmax activation.
- Concatenated Input: A line connects the input layer to each concatenated LeNet-5 model, indicating that the same input is fed into each.
- Concatenate Layer: Above the LeNet-5 blocks, there will be a block representing the concatenate layer, combining the outputs of the concatenated models.
- Reshape Layer: Above the concatenate layer, there will be a block representing the Reshape layer, reshaping the concatenated output to have dimensions (nets, 2).
- Output Layer: In the top, there will be the final output layer with shape (None, 2).
4. Results
4.1. Dataset Description
4.2. Experimentation Details
- Sensitivity (True Positive Rate): This metric highlights the model’s ability to correctly identify images with pneumonia. It is calculated as TP divided by the sum of TP and FN. A higher sensitivity indicates a stronger capability to catch actual cases of pneumonia.
- Specificity (True Negative Rate): Specificity gauges the model’s proficiency in correctly identifying non-pneumonia images. It is computed as TN divided by the sum of TN and FP. A higher specificity implies a reduced likelihood of misclassifying non-pneumonia instances.
- Precision: Defined as the ratio of true positive predictions to the total instances predicted as positive (TP/(TP + FP)), precision measures the model’s ability to accurately identify pneumonia cases among all instances it predicts as positive. In the medical diagnosis context, precision becomes crucial as it reflects the proportion of predicted positive cases that are indeed true positives. High precision indicates that when the model predicts pneumonia, it is likely to be correct, minimizing the risk of false alarms in clinical settings.
- F1-score: The F1-score combines precision and recall, striking a balance between the two metrics. It is the harmonic mean of precision and recall, providing a comprehensive measure of a model’s performance by considering both false positives and false negatives. The F1-score is particularly valuable in medical diagnosis, where achieving equilibrium between correctly identifying pneumonia cases (recall) and ensuring those predictions are accurate (precision) is paramount. A higher F1-score signifies a model that excels in both precision and recall, crucial for reliable and trustworthy pneumonia detection. Additionally, support indicates the number of actual occurrences of each class, providing context to the precision and recall values and aiding in the interpretation of the model’s performance across different class sizes.
- Feature Diversity: Each LeNet-5 model processes the input data independently, capturing different aspects and features. Concatenating these outputs likely results in a more comprehensive representation of the input data, contributing to improved accuracy.
- Parameter Sharing: Since the LeNet-5 models are identical, they share the same set of parameters. This can help in reducing the overall model complexity while still benefiting from the parallel processing of multiple instances.
- Effective Representation Learning: The architecture’s ability to achieve higher accuracy on both training and testing datasets suggests that it effectively learns and generalizes representations from the input data, outperforming other architectures like Modified LeNet, ResNet 50, and AlexNet.
- Insight into Class Imbalances: In situations where there is a class imbalance (significant difference in the number of instances between classes), a confusion matrix helps identify how well the model performs for each class.
- Model Comparison: When comparing multiple models, a confusion matrix facilitates a side-by-side evaluation of their performance, enabling stakeholders to make informed decisions about which model is better suited for a particular task.
- Diagnostic Information: The confusion matrix is particularly useful in medical and diagnostic applications, providing information on the model’s ability to correctly identify positive (disease presence) and negative (disease absence) cases.
- Ensemble Learning: The use of three identical LeNet-5 models in parallel introduced an ensemble learning strategy. Ensemble methods combine multiple models to improve overall performance by capturing diverse patterns and representations. In the concatenated LeNet-5 model, the parallel processing of three identical models enabled the extraction of complementary features from the input data. The subsequent concatenation of these diverse features contributes to a more robust and generalized model.
- Comprehensive Feature Extraction: The concatenated LeNet-5 architecture allowed for a comprehensive extraction of features from the input data. Each LeNet-5 model processed the input independently, capturing different aspects and details. By concatenating these outputs, the model can aggregate a more extensive set of features, enhancing its ability to discriminate between classes and improving accuracy.
- Parameter Sharing: The three LeNet-5 models in the concatenated LeNet-5 architecture are identical, meaning they shared the same set of parameters. Parameter sharing reduced the overall model complexity, making it more efficient and preventing overfitting on the training data. This shared parameterization also facilitates effective learning and generalization, contributing to the model’s high accuracy on both training and testing datasets.
- Reduction of Overfitting: The ensemble nature of the concatenated LeNet-5 model helped mitigate overfitting. Overfitting occurred when a model learnt to perform well on the training data but failed to generalize to unseen data. The diversity introduced by parallel LeNet-5 models and their subsequent combination through concatenation aids in reducing overfitting, leading to better generalization and higher testing accuracy.
- Parallel Processing and Efficiency: The parallel processing of three LeNet-5 models allows for efficient computation, enabling faster training times. This can be particularly advantageous when dealing with large datasets and complex architectures. The efficiency gained through parallel processing contributes to quicker convergence during training, resulting in a model that achieves higher accuracy in a shorter amount of time.
- Effective Learning Representations: The Concatenated LeNet-5 model exceled in learning effective representations of the input data. The combination of parallel processing, ensemble learning, and concatenation of outputs results in a model that captured both low-level and high-level features, contributing to its ability to make accurate predictions.
- Improved performance in confirming infected cases
- Flexibility in using different fine-tuned deep learning models
- Ability to handle multi-label classification of X-ray images
- Successful testing and evaluation on a public X-ray image dataset
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Obaro, S.K.; Madhi, S.A. Bacterial pneumonia vaccines and childhood pneumonia: Are we winning, refining, or redefining? Lancet Infect. Dis. 2006, 6, 150–161. [Google Scholar] [CrossRef]
- Pound, M.W.; Drew, R.H.; Perfect, J.R. Recent advances in the epidemiology, prevention, diagnosis, and treatment of fungal Pneumonia. Curr. Opin. Infect. Dis. 2002, 15, 183–194. [Google Scholar] [CrossRef]
- Virkki, R.; Juven, T.; Rikalainen, H.; Svedström, E.; Mertsola, J.; Ruuskanen, O. Differentiation of bacterial and viral Pneumonia in children. Thorax 2002, 57, 438–441. [Google Scholar] [CrossRef]
- Jones, R.N. Microbial etiologies of hospital-acquired bacterial Pneumonia and ventilator-associated bacterial Pneumonia. Clin. Infect. Dis. 2010, 51 (Suppl. 1), S81–S87. [Google Scholar] [CrossRef]
- Ruuskanen, O.; Lahti, E.; Jennings, L.C.; Murdoch, D.R. Viral pneumonia. Lancet 2011, 377, 1264–1275. [Google Scholar] [CrossRef]
- Sureshkumar, V.; Balasubramaniam, S.; Ravi, V.; Arunachalam, A. A hybrid optimization algorithm-based feature selection for thyroid disease classifier with rough type-2 fuzzy support vector machine. Expert Syst. 2022, 39, e12811. [Google Scholar] [CrossRef]
- World Health Organization. Revised WHO Classification and Treatment of Childhood Pneumonia at Health Facilities: Evidence Summaries; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Garg, M.; Prabhakar, N.; Kiruthika, P.; Agarwal, R.; Aggarwal, A.; Gulati, A.; Khandelwal, N. Imaging of Pneumonia: An Overview. Curr. Radiol. Rep. 2017, 5, 1–14. [Google Scholar] [CrossRef]
- Hasan, M.D.K.; Ahmed, S.; Abdullah, Z.M.E.; Khan, M.M.; Anand, D.; Singh, A.; AlZain, M.; Masud, M. Deep Learning Approaches for Detecting Pneumonia in COVID-19 Patients by Analyzing Chest X-Ray Images. Math. Probl. Eng. 2021, 2021, 1–8. [Google Scholar] [CrossRef]
- Le Cun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Fukushima, K.; Miyake, S. Neocognitron: A new algorithm for pattern recognition tolerant of deformations and shifts in position. Pattern Recognit. 1982, 15, 455–469. [Google Scholar] [CrossRef]
- Schultheiss, M.; Schmette, P.; Bodden, J.; Aichele, J.; Müller-Leisse, C.; Gassert, F.G.; Gassert, F.T.; Gawlitza, J.F.; Hofmann, F.C.; Sasse, D.; et al. Lung nodule detection in chest X-rays using synthetic ground-truth data comparing CNN-based diagnosis to human performance. Sci. Rep. 2021, 11, 15857. [Google Scholar] [CrossRef] [PubMed]
- Behrendt, F.; Bengs, M.; Bhattacharya, D.; Krüger, J.; Opfer, R.; Schlaefer, A. A systematic approach to deep learning-based nodule detection in chest radiographs. Sci. Rep. 2023, 13, 10120. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.S.; Hasikin, K.; Lai, K.W.; Zain, N.M.; Azizan, M.M. Microcalcification Discrimination in Mammography Using Deep Convolutional Neural Network: Towards Rapid and Early Breast Cancer Diagnosis. Front. Public Health 2022, 10, 875305. [Google Scholar] [CrossRef] [PubMed]
- Pesapane, F.; Trentin, C.; Ferrari, F.; Signorelli, G.; Tantrige, P.; Montesano, M.; Cicala, C.; Virgoli, R.; D’Acquisto, S.; Nicosia, L.; et al. Deep learning performance for detection and classification of microcalcifications on mammography. Eur. Radiol. Exp. 2023, 7, 69. [Google Scholar] [CrossRef]
- Stokes, K.; Castaldo, R.; Franzese, M.; Salvatore, M.; Fico, G.; Pokvic, L.G.; Badnjevic, A.; Pecchia, L. A machine learning model for supporting symptom-based referral and diagnosis of bronchitis and pneumonia in limited resource settings. Biocybern. Biomed. Eng. 2021, 41, 1288–1302. [Google Scholar] [CrossRef]
- Khaniabadi, P.M.; Bouchareb, Y.; Al-Dhuhli, H.; Shiri, I.; Al-Kindi, F.; Khaniabadi, B.M.; Zaidi, H.; Rahmim, A. Two-step machine learning to diagnose and predict involvement of lungs in COVID-19 and pneumonia using CT radiomics. Comput. Biol. Med. 2022, 150, 106165. [Google Scholar] [CrossRef]
- Kareem, A.; Liu, H.; Velisavljevic, V. A federated learning framework for pneumonia image detection using distributed data. Health Anal. 2023, 4, 100204. [Google Scholar] [CrossRef]
- Baik, S.M.; Hong, K.S.; Park, D.J. Application and utility of boosting machine learning model based on laboratory test in the differential diagnosis of non-COVID-19 pneumonia and COVID-19. Clin. Biochem. 2023, 118, 110584. [Google Scholar] [CrossRef]
- Hussain, M.A.; Mirikharaji, Z.; Momeny, M.; Marhamati, M.; Neshat, A.A.; Garbi, R.; Hamarneh, G. Active deep learning from a noisy teacher for semi-supervised 3D image segmentation: Application to COVID-19 pneumonia infection in CT. Comput. Med Imaging Graph. 2022, 102, 102127. [Google Scholar] [CrossRef]
- Sharma, S.; Guleria, K. A Deep Learning based model for the Detection of Pneumonia from Chest X-ray Images using VGG-16 and Neural Networks. Procedia Comput. Sci. 2023, 218, 357–366. [Google Scholar] [CrossRef]
- Lamia, A.; Fawaz, A. Detection of Pneumonia Infection by Using Deep Learning on a Mobile Platform. Comput. Intell. Neurosci. 2022, 2022, 7925668. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, S.; Velmurugan, Y.; Jaganathan, D.; Dhanasekaran, S. A Modified LeNet CNN for Breast Cancer Diagnosis in Ultrasound Images. Diagnostics 2023, 13, 2746. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Yun, J.; Cho, Y.; Shin, K.; Jang, R.; Bae, H.-J.; Kim, N. Deep Learning in Medical Imaging. Neurospine 2019, 16, 657–668. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Jabbar, M.K.; Yan, J.; Xu, H.; Ur Rehman, Z.; Jabbar, A. Transfer Learning-Based Model for Diabetic Retinopathy Diagnosis Using Retinal Images. Brain Sci. 2022, 12, 535. [Google Scholar] [CrossRef]
- Pan, S.J.; Yang, Q. A Survey on Deep Transfer Learning for Image Classification. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Reenadevi, R.; Sathiyabhama, B.; Vinayakumar, R.; Sankar, S. Hybrid Optimization Algorithm based feature selection for mammogram images and detecting the breast mass using Multilayer Perceptron classifier. J. Comput. Intell. 2022, 38, 1559–1593. [Google Scholar]
- Tan, Y.N.; Tinh, V.P.; Lam, P.D.; Nam, N.H.; Khoa, T.A. A Transfer Learning Approach to Breast Cancer Classification in a Federated Learning Framework. IEEE Access 2023, 11, 27462–27476. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.U.; Ozsoz, M.; Serte, S.; Al-Turjman, F.; Yakoi, P.S. Pneumonia Classification Using Deep Learning from Chest X-ray Images During COVID-19. Cogn. Comput. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kundu, R.; Das, R.; Geem, Z.W.; Hen, G.-T.; Sarkar, R. Pneumonia Detection in Chest X-ray images using an Ensemble of Deep Learning Models. PLoS ONE 2021, 16, e0256630. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J. Based on improved deep convolutional neural network model pneumonia image classification. PLoS ONE 2021, 16, e0258804. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, X.; Farrahi, K.; Niranjan, M. Deep Cascade Learning for Optimal Medical Image Feature Representation. Proc. Mach. Learn. Res. Mach. Learn. Healthc. 2022, 182, 1–25. [Google Scholar]
- Zhang, Y. Cascade of Classifier Ensembles for Reliable Medical Image Classification. Ph.D. Thesis, University of Liverpool, Liverpool, UK, 2014. [Google Scholar] [CrossRef]
- Aljawarneh, S.A.; Al-Quraan, R. Pneumonia Detection Using Enhanced Convolutional Neural Network Model on Chest X-Ray Images. Big Data 2023. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-F.; Cheng, H.-T.; Wei, A.; Chen, H.-M.; Kuo, P.-C.; Liu, K.-C.; Ko, M.-C.; Chen, R.-J.; Lee, P.-C.; Chuang, J.-H.; et al. A Cascaded Learning Strategy for Robust COVID-19 Pneumonia Chest X-Ray Screening. arXiv 2020, arXiv:2004.12786v2. [Google Scholar]
- Karar, M.E.; Hemdan, E.E.; Shouman, M.A. Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Complex Intell. Syst. 2021, 7, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.; Zhang, K.; Goldbaum, M. Labeled Optical Coherence Tomography (OCT) and Chest X-ray Images for Classification. Mendeley Data 2018, 2, 651. [Google Scholar] [CrossRef]
- Vidhushavarshini, S.; Sathiyabhama, B. A Comparison of Classification Techniques on Thyroid Detection Using J48 and Naive Bayes Classification Techniques. In Proceedings of the International Conference on Intelligent Computing Systems (ICICS 2017), Salem, Tamilnadu, India, 15–16 December 2017. [Google Scholar]
- Mei, X.; Liu, Z.; Singh, A.; Lange, M.; Boddu, P.; Gong, J.Q.X.; Lee, J.; DeMarco, C.; Cao, C.; Platt, S.; et al. Interstitial lung disease diagnosis and prognosis using an AI system integrating longitudinal data. Nat. Commun. 2023, 14, 1–11. [Google Scholar] [CrossRef]
- Chest X-ray Images. Available online: https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia (accessed on 3 May 2023).












| Class Prediction | Precision | Recall | f1-Score | Support |
|---|---|---|---|---|
| Non-Pneumonia | 0.93 | 0.90 | 0.92 | 306 |
| Pneumonia | 0.97 | 0.98 | 0.97 | 866 |
| Accuracy | 0.96 | 1172 | ||
| weighted avg | 0.95 | 0.94 | 0.94 | 1172 |
| weighted avg | 0.96 | 0.96 | 0.96 | 1172 |
| Predicted | |||
|---|---|---|---|
| 0 | 1 | ||
| Actual | 0 | True Negative 293 | False Positive 22 |
| 1 | False Negative 33 | True Table 2 Positive 824 | |
| Training Accuracy | Test Accuracy | |
|---|---|---|
| Concatenated LeNet | 99 | 96 |
| Modified LeNet | 89 | 87 |
| Resnet50 | 85 | 84 |
| Alexnet | 81 | 79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaganathan, D.; Balsubramaniam, S.; Sureshkumar, V.; Dhanasekaran, S. Concatenated Modified LeNet Approach for Classifying Pneumonia Images. J. Pers. Med. 2024, 14, 328. https://doi.org/10.3390/jpm14030328
Jaganathan D, Balsubramaniam S, Sureshkumar V, Dhanasekaran S. Concatenated Modified LeNet Approach for Classifying Pneumonia Images. Journal of Personalized Medicine. 2024; 14(3):328. https://doi.org/10.3390/jpm14030328
Chicago/Turabian StyleJaganathan, Dhayanithi, Sathiyabhama Balsubramaniam, Vidhushavarshini Sureshkumar, and Seshathiri Dhanasekaran. 2024. "Concatenated Modified LeNet Approach for Classifying Pneumonia Images" Journal of Personalized Medicine 14, no. 3: 328. https://doi.org/10.3390/jpm14030328
APA StyleJaganathan, D., Balsubramaniam, S., Sureshkumar, V., & Dhanasekaran, S. (2024). Concatenated Modified LeNet Approach for Classifying Pneumonia Images. Journal of Personalized Medicine, 14(3), 328. https://doi.org/10.3390/jpm14030328