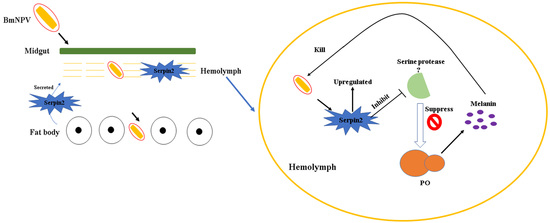
Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rearing of Silkworm and B. mori Nucleopolyhedrovirus (BmNPV) Preparation
2.2. BmNPV Infection to B. mori
2.3. Total RNA Isolation and Amplification of BmSerpin2
2.4. Recombinant Bmserpin2 and Antibody Preparation
2.5. Preparation of Soluble Bmserpin2
2.6. Expression Analysis of Bmserpin2 in Different Tissues and Developmental Stages
2.7. Expression Analysis of Bmserpin2 after BmNPV Infection
2.8. Western Blotting
2.9. Effect of Bmserpin2 on the Melanization of Haemolymph
2.10. Synthesis and Micro-Injection of Small Interfering RNA (siRNA)
2.11. Phenoloxidase (PO) Activity Assay
2.12. Examination of Viral DNA in Haemolymph
3. Results
3.1. Tissue Distribution and Expression of BmSerpin2 Gene at Different Developmental Stages
3.2. Expression Analysis of Bmserpin2 in Response to BmNPV Infection
3.3. Bmserpin2 Inhibits Melanization in Haemolymph
3.4. Silencing of Bmserpin2 in B. mori Increased PO Activity in Response to BmNPV Infection
3.5. Virus Genomic DNA Copies Decreased in BmSerpin2-Depleted Haemolymph
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus diversity and molecular biology. Ann. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Li, M.W.; Wang, Y.; Wang, W.B.; Lu, J.; Dong, Y.; Chen, K.P. Screening of molecular markers for NPV resistance in Bombyx mori L. (Lep., Bombycidae). J. Appl. Entomol. 2003, 127, 134–136. [Google Scholar] [CrossRef]
- Bao, Y.-Y.; Tang, X.-D.; Lv, Z.-Y.; Wang, X.-Y.; Tian, C.-H.; Xu, Y.P.; Zhang, C.-X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Lavine, M.D.; Strand, M.R. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The Host defense of drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]
- Steinert, S.; Levashina, E.A. Intracellular immune responses of dipteran insects. Immunol. Rev. 2011, 240, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, Y.; Yu, X.-Q.; Zhu, Y.; Kanost, M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: A clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 2003, 33, 1049–1060. [Google Scholar] [CrossRef]
- Kanost, M.R.; Gorman, M.J. 4–phenoloxidases in insect immunity. In Insect Immunology; Beckage, N.E., Ed.; Academic Press: San Diego, CA, USA, 2008; pp. 69–96. [Google Scholar] [CrossRef]
- Lu, A.; Zhang, Q.; Zhang, J.; Yang, B.; Wu, K.; Xie, W.; Luan, Y.X.; Ling, E. Insect prophenoloxidase: The view beyond immunity. Front. Physiol. 2014, 5, 252. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999, 23, 291–301. [Google Scholar] [CrossRef]
- Gettins, P.G. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011, 9, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Kanost, M.R.; Michel, K. Serpins in arthropod biology. Semin. Cell Dev. Biol. 2017, 62, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Garrett, M.; Fullaondo, A.; Troxler, L.; Micklem, G.; Gubb, D. Identification and analysis of serpin-family genes by homology and synteny across the 12 sequenced Drosophilid genomes. BMC Genom. 2009, 10, 489. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Picheng, Z.; Weng, H.; Mita, K.; Jiang, H. A comparative analysis of serpin genes in the silkworm genome. Genomics 2009, 93, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Christen, J.M.; Dittmer, N.T.; Cao, X.; Zhang, X.; Jiang, H.; Kanost, M.R. The Manduca sexta serpinome: Analysis of serpin genes and proteins in the tobacco hornworm. Insect Biochem. Mol. Biol. 2018, 102, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.H.; Xing, L.S.; Lin, Z.; Saha, T.T.; Wang, C.; Jiang, H.; Zou, Z. High throughput profiling of the cotton bollworm Helicoverpa armigera immunotranscriptome during the fungal and bacterial infections. BMC Genomics 2015, 16, 321. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Zhang, B.; Kurokawa, K.; So, Y.-I.; Kim, E.-H.; Hwang, H.O.; Lee, J.-H.; Shiratsuchi, A.; Zhang, J.; Nakanishi, Y.; et al. 93-kDa twin-domain serine protease inhibitor (serpin) has a regulatory function on the beetle toll proteolytic signaling cascade. J. Biol. Chem. 2011, 286, 35087–35095. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Evans, J.D.; Lu, Z.; Zhao, P.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007, 8, R177. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Y.; Gorman, M.J.; Jiang, H.; Kanost, M.R. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J. Biol. Chem. 2003, 278, 46556–46564. [Google Scholar] [CrossRef] [PubMed]
- Suwanchaichinda, C.; Ochieng, R.; Zhuang, S.; Kanost, M.R. Manduca sexta serpin-7, a putative regulator of hemolymph prophenoloxidase activation. Insect Biochem. Mol. Biol. 2013, 43, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, Y.; Sumathipala, N.; Cao, X.; Kanost, M.R.; Jiang, H. Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph. Insect Biochem. Mol. Biol. 2018, 99, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, L.; Lin, Z.; Zou, Z.; Lu, Z. Serpin-5 regulates prophenoloxidase activation and antimicrobial peptide pathways in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 73, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yu, H.-Z.; Ye, C.-J.; Ma, Y.; Li, X.; Fan, T.; Chen, F.-S.; Xu, J.-P. Bombyx mori Serpin6 regulates prophenoloxidase activity and the expression of antimicrobial proteins. Gene 2017, 610, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, L.; Yang, L.; Qian, C.; Wei, G.; Dai, L.; Li, J.; Zhu, B.; Liu, C. Serpin-15 from Bombyx mori inhibits prophenoloxidase activation and expression of antimicrobial peptides. Dev. Comp. Immunol. 2015, 51, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.-C.; Dong, Z.; Zhao, P.; Zhang, Y.; He, H.; Tan, X.; Zhang, W.; Xia, Q. Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18. Sci. Rep. 2015, 5, 11863. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H.; Fu, H.; Zhang, L.; Guo, P.; Xia, Q.; Zhao, P. Silkworm serpin32 functions as a negative-regulator in prophenoloxidase activation. Dev. Comp. Immunol. 2019, 91, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Shelby, K.S.; Popham, H.J. Plasma phenoloxidase of the larval tobacco budworm, Heliothis virescens, is virucidal. J. Insect Sci. (Online) 2006, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Lu, Z.; Strand, M.R.; Jiang, H. Antiviral, anti-parasitic, and cytotoxic effects of 5,6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochem. Mol. Biol. 2011, 41, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Andres, J.; Rani, S.; Varjak, M.; Chase-Topping, M.E.; Beck, M.H.; Ferguson, M.C.; Schnettler, E.; Fragkoudis, R.; Barry, G.; Merits, A.; et al. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 2012, 8, e1002977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, C.; Xing, L.; Wang, M.; Wang, X.; Yin, M.; Wang, Q.; Hu, Z.; Zou, Z. Inhibition of melanization by serpin-5 and serpin-9 promotes baculovirus infection in cotton bollworm Helicoverpa armigera. PLoS Pathog. 2017, 13, e1006645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.-Y.; Yu, H.-Z.; Geng, L.; Xu, J.-P.; Yu, D.; Zhang, S.-Z.; Ma, Y.; Fei, D.-Q. Comparative transcriptome analysis of Bombyx mori (Lepidoptera) larval midgut response to bmnpv in susceptible and near-isogenic resistant strains. PLoS ONE 2016, 11, e0155341. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Gopinathan, K.P. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 2004, 101, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, X.; Xu, J.; Ma, Y.; Zhang, S.; Yu, D.; Fei, D.; Muhammad, A. iTRAQ-based quantitative proteomics analysis of molecular mechanisms associated with Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic strains. J. Proteom. 2017, 165, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, T.; Zhan, M.-Y.; Yang, P.-J.; Yu, X.-Q.; Rao, X.-J. Molecular cloning and analysis of a C-type lectin from silkworm Bombyx mori. Arch. Insect Biochem. Physiol. 2017, 95, e21391. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yao, L.; Zhang, J.; Huang, L. Direct and indirect effects of RNA interference against pyridoxal kinase and pyridoxine 5′-phosphate oxidase genes in Bombyx mori. Gene 2016, 587, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.Y.; Lv, Z.Y.; Liu, Z.B.; Xue, J.; Xu, Y.P.; Zhang, C.X. Comparative analysis of Bombyx mori nucleopolyhedrovirus responsive genes in fat body and haemocyte of B. mori resistant and susceptible strains. Insect Mol. Biol. 2010, 19, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Kausar, S.; Abbas, M.N.; Qian, C.; Zhu, B.; Sun, Y.; Sun, Y.; Wang, L.; Wei, G.; Maqsood, I.; Liu, C.-L. Serpin-14 negatively regulates prophenoloxidase activation and expression of antimicrobial peptides in Chinese oak silkworm Antheraea pernyi. Dev. Comp. Immunol. 2017, 76, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ferrandon, D.; Imler, J.-L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, L.; Dai, J.; Yuan, G.; Wang, L.; Qian, C.; Zhu, B.; Liu, C.; Wei, G. Characterization and functional analysis of serpin-28 gene from silkworm, Bombyx mori. J. Invertebr. Pathol. 2018, 159, 18–27. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Kanost, M.R. Manduca sexta serpin-5 regulates prophenoloxidase activation and the Toll signaling pathway by inhibiting hemolymph proteinase HP6. Insect Biochem. Mol. Biol. 2010, 40, 683–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, Y.; Zhou, F.; Liu, Y.; Hong, F.; Wang, G.; An, C. Ostrinia furnacalis serpin-3 regulates melanization cascade by inhibiting a prophenoloxidase-activating protease. Insect Biochem. Mol. Biol. 2015, 61, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.D. Altered tyrosine metabolism and melanization complex formation underlie the developmental regulation of melanization in Manduca sexta. Insect Biochem. Mol. Biol. 2015, 58, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Sangsuriya, P.; Charoensapsri, W.; Sutthangkul, J.; Senapin, S.; Hirono, I.; Tassanakajon, A.; Amparyup, P. A novel white spot syndrome virus protein WSSV164 controls prophenoloxidases, PmproPOs in shrimp melanization cascade. Dev. Comp. Immunol. 2018, 86, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Beck, M.H.; Wang, Y.; Jiang, H.; Strand, M.R. The viral protein Egf1.0 is a dual activity inhibitor of prophenoloxidase-activating proteinases 1 and 3 from Manduca sexta. J. Biol. Chem. 2008, 283, 21325–21333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Beck, M.H.; Strand, M.R. Egf1.5 is a second phenoloxidase cascade inhibitor encoded by Microplitis demolitor bracovirus. Insect Biochem. Mol. Biol. 2010, 40, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-T.; Hu, N.; Tan, L.-R.; Xiao, Q.; Dong, Z.-Q.; Chen, P.; Xu, A.-Y.; Pan, M.-H.; Lu, C. Resistant silkworm strain block viral infection independent of melanization. Pestic. Biochem. Physiol. 2019, 154, 88–96. [Google Scholar] [CrossRef] [PubMed]











| Primers | Sequences of Primers (5′ to 3′) | Purpose |
|---|---|---|
| Bmserpin2-F | CGCGGATCCATGATATTTGCCAGAGTTTTGTT | Protein expression |
| Bmserpin2-R | CCGCTCGAGTCAATTTCGTCCACGGTATT | Protein expression |
| Bmserpin2-F | CAGAATACTTGCTGGCGTTGATA | qRT-PCR |
| Bmserpin2-R | AGCCGAGAATGATGAGCGAAT | qRT-PCR |
| BmGAPDH-F | CATTCCGCGTCCCTGTTGCTAAT | qRT-PCR |
| BmGAPDH-R | GCTGCCTCCTTGACCTTTTGC | qRT-PCR |
| GP64-F | GAAGTAGAAACGCCGCATCG | qRT-PCR |
| GP64-R | GTGGGGTATTCAGGCAGCAGT | qRT-PCR |
| Name | Sense Strand | Antisense Strand |
|---|---|---|
| siRNA-1 | GGAAAUAAAGAACCCUGUUTT | AACAGGGUUCUUUAUUUCCTT |
| siRNA-2 | GCUGCCGAACGAAAUUAAUTT | AUUAAUUUCGUUCGGCAGCTT |
| siGFP | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toufeeq, S.; Wang, J.; Zhang, S.-Z.; Li, B.; Hu, P.; Zhu, L.-B.; You, L.-L.; Xu, J.-P. Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori. Insects 2019, 10, 399. https://doi.org/10.3390/insects10110399
Toufeeq S, Wang J, Zhang S-Z, Li B, Hu P, Zhu L-B, You L-L, Xu J-P. Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori. Insects. 2019; 10(11):399. https://doi.org/10.3390/insects10110399
Chicago/Turabian StyleToufeeq, Shahzad, Jie Wang, Shang-Zhi Zhang, Bing Li, Pei Hu, Lin-Bao Zhu, Ling-Ling You, and Jia-Ping Xu. 2019. "Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori" Insects 10, no. 11: 399. https://doi.org/10.3390/insects10110399
APA StyleToufeeq, S., Wang, J., Zhang, S. -Z., Li, B., Hu, P., Zhu, L. -B., You, L. -L., & Xu, J. -P. (2019). Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori. Insects, 10(11), 399. https://doi.org/10.3390/insects10110399