Microbiome Associated with the Mycangia of Female and Male Adults of the Ambrosia Beetle Platypus cylindrus Fab. (Coleoptera: Curculionidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Beetle Mycangia Sampling
2.2. DNA Extraction and Sequencing
2.3. Bioinformatic Analysis
2.4. Statistical Analysis
2.5. Analysis of Mycangia Anatomical Structure
3. Results
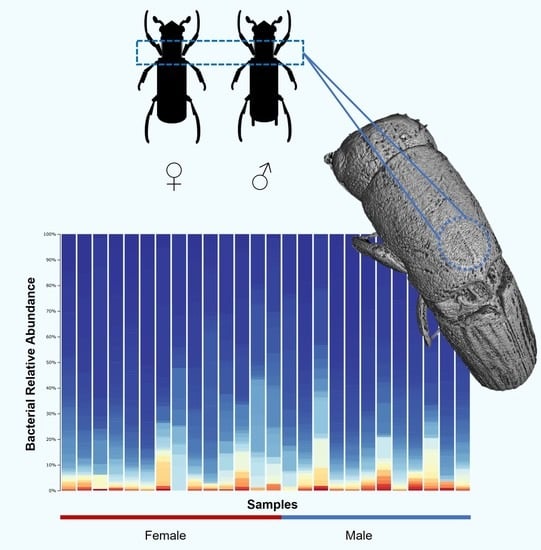
3.1. Bacterial Community Composition
3.2. Core Bacteriome
3.3. Microbial Population Diversity
3.4. Anatomical Structure of Mycangia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hulcr, J.; Stelinski, L.L. The Ambrosia Symbiosis: From Evolutionary Ecology to Practical Management. Annu. Rev. Entomol. 2017, 62, 285–303. [Google Scholar] [CrossRef] [Green Version]
- Vanderpool, D.; Bracewell, R.R.; McCutcheon, J.P. Know Your Farmer: Ancient Origins and Multiple Independent Domestications of Ambrosia Beetle Fungal Cultivars. Mol. Ecol. 2018, 27, 2077–2094. [Google Scholar] [CrossRef]
- Hulcr, J.; Rountree, N.R.; Diamond, S.E.; Stelinski, L.L.; Fierer, N.; Dunn, R.R. Mycangia of Ambrosia Beetles Host Communities of Bacteria. Microb. Ecol. 2012, 64, 784–793. [Google Scholar] [CrossRef] [Green Version]
- Cassier, P.; Lévieux, J.; Morelet, M.; Rougon, D. The Mycangia of Platypus cylindrus Fab. and P. oxyurus Dufour (Coleoptera: Platypodidae). Structure and Associated Fungi. J. Insect Physiol. 1996, 42, 171–179. [Google Scholar] [CrossRef]
- Scott, J.J.; Oh, D.-C.; Yuceer, M.C.; Klepzig, K.D.; Clardy, J.; Currie, C.R. Bacterial Protection of Beetle-Fungus Mutualism. Science 2008, 322, 63. [Google Scholar] [CrossRef] [Green Version]
- Ibarra-Juarez, L.; Desgarennes, D.; Vázquez-Rosas-Landa, M.; Villafan, E.; Alonso-Sánchez, A.; Ferrera-Rodríguez, O.; Moya, A.; Carrillo, D.; Cruz, L.; Carrión, G.; et al. Impact of Rearing Conditions on the Ambrosia Beetle’s Microbiome. Life 2018, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarra-Juarez, L.A.; Burton, M.A.J.; Biedermann, P.H.W.; Cruz, L.; Desgarennes, D.; Ibarra-Laclette, E.; Latorre, A.; Alonso-Sánchez, A.; Villafan, E.; Hanako-Rosas, G.; et al. Evidence for Succession and Putative Metabolic Roles of Fungi and Bacteria in the Farming Mutualism of the Ambrosia Beetle Xyleborus affinis. mSystems 2020, 5, e00541-20. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, C.; Chen, H.; Ma, J. Differences in the Structure of the Gut Bacteria Communities in Development Stages of the Chinese White Pine Beetle (Dendroctonus armandi). Int. J. Mol. Sci. 2013, 14, 21006–21020. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lu, M.; Xu, D.; Chen, L.; Sun, J. Sexual Variation of Bacterial Microbiota of Dendroctonus valens Guts and Frass in Relation to Verbenone Production. J. Insect Physiol. 2016, 95, 110–117. [Google Scholar] [CrossRef]
- Sousa, E.; Inácio, M.L. New aspects of Platypus cylindrus Fab. (Coleoptera: Platypodidae) life history on cork oak stands in Portugal. In Entomological Research in Mediterranean Forest Ecosystems; Lieutier, F., Ghaioule, D., Eds.; INRA Editions: Paris, France, 2005; pp. 147–168. [Google Scholar]
- Inácio, M.L.; Henriques, J.; Sousa, E. Contribution of Symbiotic Fungi to Cork Oak Colonization by Platypus cylindrus (Coleoptera: Platypodidae). Silva Lusit. 2011, 19, 89–99. [Google Scholar]
- Inácio, M.L.; Henriques, J.; Lima, A.; Sousa, E. Ophiostomatoid Fungi Associated with Cork Oak Mortality in Portugal. IOBC Wprs Bull. 2012, 76, 89–92. [Google Scholar]
- Nones, S.; Cruz, L.; Fernandes, C.; Duarte, L.; Sousa, E. New Insights into the Bacterial Diversity Associated with Symptomatic Quercus suber Trees Infested by the Ambrosia Beetle Platypus cylindrus (Coleoptera: Curculionidae). IOBC Wprs Bull. 2020, 152, 51–54. [Google Scholar]
- Tarno, H.; Septia, E.D.; Aini, L.Q. Microbial Community Associated with Ambrosia Beetle, Euplatypus parallelus on Sonokembang, Pterocarpus indicus in Malang. AGRIVITA J. Agric. Sci. 2016, 38, 312–320. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference; SciPy: Austin, TX, USA, 2010; Volume 445, pp. 51–56. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Vázquez-Baeza, Y.; Koslicki, D.; McClelland, J.; Reeve, N.; Xu, Z.; Gonzalez, A.; Knight, R. Striped UniFrac: Enabling Microbiome Analysis at Unprecedented Scale. Nat. Methods 2018, 15, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Robeson, M. Make SILVA Database: General Procedure for Making QIIME 2 Compatible SILVA Reference Files. Available online: https://github.com/mikerobeson/make_SILVA_db (accessed on 1 August 2020).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Vanderplas, J. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 122825–122830. [Google Scholar]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Sapp, M.; Lewis, E.; Moss, S.; Barrett, B.; Kirk, S.; Elphinstone, J.; Denman, S. Metabarcoding of Bacteria Associated with the Acute Oak Decline Syndrome in England. Forests 2016, 7, 95. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [Green Version]
- Chang, Q.; Luan, Y.; Sun, F. Variance Adjusted Weighted UniFrac: A Powerful Beta Diversity Measure for Comparing Communities Based on Phylogeny. BMC Bioinform. 2011, 12, 118. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating Microbiome Composition with Environmental Covariates Using Generalized UniFrac Distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A Tool for Visualizing High-Throughput Microbial Community Data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Russel, J.; Thorsen, J.; Brejnrod, A.D.; Bisgaard, H.; Sørensen, S.J.; Burmølle, M. DAtest: A Framework for Choosing Differential Abundance or Expression Method. BioRxiv 2018, 241802. [Google Scholar] [CrossRef] [Green Version]
- Johansen, D.A. Plant Microtechnique; McGraw-Hill Publishing Company Ltd.: London, UK, 1940. [Google Scholar]
- Ruzin, S.E. Plant Microtechnique and Microscopy; Oxford University Press: Oxford, NY, USA, 1999; ISBN 978-0-19-508956-1. [Google Scholar]
- Spurr, A.R. A Low-Viscosity Epoxy Resin Embedding Medium for Electron Microscopy. J. Ultrastruct. Res. 1969, 26, 31–43. [Google Scholar] [CrossRef]
- Cardoza, Y.J.; Vasanthakumar, A.; Suazo, A.; Raffa, K.F. Survey and Phylogenetic Analysis of Culturable Microbes in the Oral Secretions of Three Bark Beetle Species. Entomol. Exp. Appl. 2009, 131, 138–147. [Google Scholar] [CrossRef]
- Kati, A.; Kati, H. Isolation and Identification of Bacteria from Xylosandrus germanus (Blandford) (Coleoptera: Curculionidae). Afr. J. Microbiol. Res. 2013, 7, 5288–5299. [Google Scholar] [CrossRef]
- Vicente, C.S.L.; Nascimento, F.X.; Espada, M.; Barbosa, P.; Hasegawa, K.; Mota, M.; Oliveira, S. Characterization of Bacterial Communities Associated with the Pine Sawyer Beetle Monochamus galloprovincialis, the Insect Vector of the Pinewood Nematode Bursaphelenchus xylophilus. FEMS Microbiol. Lett. 2013, 347, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Hernández-García, J.A.; Briones-Roblero, C.I.; Rivera-Orduña, F.N.; Zúñiga, G. Revealing the Gut Bacteriome of Dendroctonus Bark Beetles (Curculionidae: Scolytinae): Diversity, Core Members and Co-Evolutionary Patterns. Sci. Rep. 2017, 7, 13864. [Google Scholar] [CrossRef]
- Vďačný, P.; Érseková, E.; Šoltys, K.; Budiš, J.; Pecina, L.; Rurik, I. Co-Existence of Multiple Bacterivorous Clevelandellid Ciliate Species in Hindgut of Wood-Feeding Cockroaches in Light of Their Prokaryotic Consortium. Sci. Rep. 2018, 8, 17749. [Google Scholar] [CrossRef]
- Li, H.; Wu, S.; Wirth, S.; Hao, Y.; Wang, W.; Zou, H.; Li, W.; Wang, G. Diversity and Activity of Cellulolytic Bacteria, Isolated from the Gut Contents of Grass Carp (Ctenopharyngodon idellus) (Valenciennes) Fed on Sudan Grass (Sorghum sudanense) or Artificial Feedstuffs. Aquac. Res. 2016, 47, 153–164. [Google Scholar] [CrossRef]
- Premalatha, N.; Gopal, N.O.; Jose, P.A.; Anandham, R.; Kwon, S.-W. Optimization of Cellulase Production by Enhydrobacter sp. ACCA2 and Its Application in Biomass Saccharification. Front. Microbiol. 2015, 6, 1046. [Google Scholar] [CrossRef] [Green Version]
- Boudiaf, I.; Le Roux, C.; Baudoin, E.; Galiana, A.; Beddiar, A.; Prin, Y.; Duponnois, R. Soil Bradyrhizobium Population Response to Invasion of a Natural Quercus suber Forest by the Introduced Nitrogen-Fixing Tree Acacia mearnsii in El Kala National Park, Algeria. Soil Biol. Biochem. 2014, 70, 162–165. [Google Scholar] [CrossRef]
- Shan, S.; Wang, W.; Song, C.; Wang, M.; Sun, B.; Li, Y.; Fu, Y.; Gu, X.; Ruan, W.; Rasmann, S. The Symbiotic Bacteria Alcaligenes faecalis of the Entomopathogenic Nematodes Oscheius spp. Exhibit Potential Biocontrol of Plant- and Entomopathogenic Fungi. Microb. Biotechnol. 2019, 12, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tláskal, V.; Zrůstová, P.; Vrška, T.; Baldrian, P. Bacteria Associated with Decomposing Dead Wood in a Natural Temperate Forest. FEMS Microbiol. Ecol. 2017, 93, 93. [Google Scholar] [CrossRef]
- Baldrian, P. Forest Microbiome: Diversity, Complexity and Dynamics. FEMS Microbiol. Rev. 2016, 41, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Kastman, E.K.; Kamelamela, N.; Norville, J.W.; Cosetta, C.M.; Dutton, R.J.; Wolfe, B.E. Biotic Interactions Shape the Ecological Distributions of Staphylococcus Species. MBio 2016, 7, e01157-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGonigle, J.E.; Purves, J.; Rolff, J. Intracellular Survival of Staphylococcus aureus during Persistent Infection in the Insect Tenebrio molitor. Dev. Comp. Immunol. 2016, 59, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Alijani, Z.; Amini, J.; Ashengroph, M.; Bahramnejad, B. Antifungal Activity of Volatile Compounds Produced by Staphylococcus sciuri Strain MarR44 and Its Potential for the Biocontrol of Colletotrichum nymphaeae, Causal Agent Strawberry Anthracnose. Int. J. Food Microbiol. 2019, 307, 108276. [Google Scholar] [CrossRef] [PubMed]
- Henriques, J.; de Lurdes Inácio, M.; Sousa, E. Ambrosia Fungi in the Insect-Fungi Symbiosis in Relation to Cork Oak Decline. Rev. Iberoam. Micol. 2006, 23, 185–188. [Google Scholar] [CrossRef]
- Costa, R.; Lourenço, A.; Oliveira, V.; Pereira, H. Chemical Characterization of Cork, Phloem and Wood from Different Quercus suber Provenances and Trees. Heliyon 2019, 5, e02910. [Google Scholar] [CrossRef] [Green Version]
- De las Rivas, B.; Rodríguez, H.; Anguita, J.; Muñoz, R. Bacterial Tannases: Classification and Biochemical Properties. Appl. Microbiol. Biotechnol. 2019, 103, 603–623. [Google Scholar] [CrossRef] [Green Version]
- Sallé, A.; Nageleisen, L.-M.; Lieutier, F. Bark and Wood Boring Insects Involved in Oak Declines in Europe: Current Knowledge and Future Prospects in a Context of Climate Change. For. Ecol. Manag. 2014, 328, 79–93. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, Q.; Chen, L.; Carballar-Lejarazú, R.; Zhang, A.; Shao, E.; Liang, G.; Hu, X.; Wang, R.; Xu, L.; et al. Characterization of Bacterial Communities Associated with the Pinewood Nematode Insect Vector Monochamus alternatus Hope and the Host Tree Pinus massoniana. BMC Genom. 2020, 21, 337. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Schumann, P.; Prauser, H. The Family Cellulomonadaceae. Prokaryotes 2006, 3, 983–1001. [Google Scholar] [CrossRef]
- Wang, C.; Guo, X.; Deng, H.; Dong, D.; Tu, Q.; Wu, W. New Insights into the Structure and Dynamics of Actinomycetal Community during Manure Composting. Appl. Microbiol. Biotechnol. 2014, 98, 3327–3337. [Google Scholar] [CrossRef]
- Belhoucine, L.; Bouhraoua, R.T.; Prats, E.; Pulade-Villar, J. Fine Structure and Functional Comments of Mouthparts in Platypus cylindrus (Col., Curculionidae: Platypodinae). Micron 2013, 45, 74–82. [Google Scholar] [CrossRef]
- Six, D.L. Ecological and Evolutionary Determinants of Bark Beetle—Fungus Symbioses. Insects 2012, 3, 339–366. [Google Scholar] [CrossRef] [Green Version]
- Spahr, E.; Kasson, M.T.; Kijimoto, T. Micro-Computed Tomography Permits Enhanced Visualization of Mycangia across Development and between Sexes in Euwallacea Ambrosia Beetles. PLoS ONE 2020, 15, e0236653. [Google Scholar] [CrossRef] [PubMed]
- Moon, M.-J.; Park, J.-G.; Seo, S.-T.; Kim, K.-H. Microstructure of the Prothoracic Mycangia in the Ambrosia Beetle Platypus koryoensis (Coleoptera: Curculionidae: Platypodinae). J. Asia-Pac. Entomol. 2012, 15, 51–57. [Google Scholar] [CrossRef]
- Kent, D.S. Mycangia of the Ambrosia Beetle, Austroplatypus incompertus (Schedl) (Coleoptera: Curculionidae: Platypodinae). Aust. J. Entomol. 2008, 47, 9–12. [Google Scholar] [CrossRef]
- Nakashima, T. Notes on the Mycetangia of the Ambrosia Beetles, Platypus severini BLANDFORD and P. calamus BLANDFORD(Coleoptera: Platypodidae). Appl. Entomol. Zool. 1972, 7, 217–225. [Google Scholar] [CrossRef]
- Nakashima, T. Several Types of the Mycetangia Found in Platypodid Ambrosia Beetles (Coleoptera: Platypodidae). Insecta matsumurana. New Ser. J. Fac. Agric. Hokkaido Univ. 1975, 7, 1–69. [Google Scholar]
- Kudo, R.; Masuya, H.; Endoh, R.; Kikuchi, T.; Ikeda, H. Gut Bacterial and Fungal Communities in Ground-Dwelling Beetles Are Associated with Host Food Habit and Habitat. ISME J. 2019, 13, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Nones, S.; Simões, F.; Trindade, C.S.; Matos, J.; Sousa, E. Bacterial Communities Associated with Platypus cylindrus Fab. (Coleoptera: Curculionidae). Available online: doi.org/10.5281/zenodo.4899899 (accessed on 4 June 2021).







| Phylum | Genus | Female Mycangia (%) | Male Mycangia (%) | ||
|---|---|---|---|---|---|
| Actinobacteriota | Dermacoccaceae | 8.26% | 1.53% | ||
| Gryllotalpicola | 3.28% | 0.21% | |||
| Corynebacterium | 2.95% | 8.58% | |||
| Corynebacteriaceae | 2.64% | 0.42% | |||
| Microbacteriaceae | 1.70% | ‡ | 0.21% | ||
| Cellulomonadaceae | 1.18% | 0.08% | |||
| Nocardioides | 1.10% | 0.85% | |||
| Flexivirga | 1.04% | 0.05% | |||
| Actinomyces | 0.81% | 2.86% | |||
| Pseudoclavibacter | 0.77% | †/‡ | 0.05% | ||
| Lawsonella | 0.56% | 0.28% | |||
| Intrasporangiaceae | 0.43% | 0.52% | |||
| Rothia | 0.26% | 1.24% | †/‡ | ||
| Bacteroidota | Hydrotalea | 6.15% | 9.37% | ||
| Sediminibacterium | 6.01% | 8.43% | |||
| Arachidicoccus | 0.85% | 0.15% | |||
| Chitinophagaceae | 0.64% | † | 0.09% | ||
| Firmicutes | Staphylococcus | 2.54% | 1.72% | ||
| Streptococcus | 2.48% | 7.53% | |||
| Anaerococcus | 0.25% | 0.58% | |||
| Lactobacillales | 0.16% | 0.98% | †/‡ | ||
| Fusobacteriota | Leptotrichia | 0.02% | 1.17% | †/‡ | |
| Proteobacteria | Enterobacterales | 7.53% | 1.69% | ||
| Pseudoxanthomonas | 3.98% | 0.45% | |||
| Pseudomonas | 3.48% | 0.35% | |||
| Bradyrhizobium | 3.07% | 4.30% | |||
| Alcaligenes | 2.50% | 4.14% | |||
| Sphingomonas | 2.36% | 2.56% | |||
| Comamonadaceae | 1.40% | 1.77% | |||
| Shimwellia | 1.38% | 1.74% | |||
| Neisseriaceae uncultured | 1.20% | 0.77% | |||
| Sphingomonadaceae | 1.02% | 1.69% | ‡ | ||
| Endobacter | 0.97% | 0.46% | |||
| Izhakiella | 0.92% | 0.04% | |||
| Phyllobacterium | 0.81% | 0.87% | |||
| Enterobacteriaceae | 0.77% | ‡ | 0.45% | ||
| Erwiniaceae | 0.67% | † | 0.04% | ||
| Enhydrobacter | 0.66% | 1.84% | |||
| Acinetobacter | 0.63% | ‡ | 1.04% | ||
| Undibacterium | 0.58% | 0.97% | |||
| Labrys | 0.52% | 0.00% | |||
| Pannonibacter | 0.49% | 0.63% | |||
| Ochrobactrum | 0.35% | 0.50% | |||
| Rhodobacteraceae | 0.30% | 0.55% | |||
| Neisseria | 0.14% | 1.05% | †/‡ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nones, S.; Simões, F.; Trindade, C.S.; Matos, J.; Sousa, E. Microbiome Associated with the Mycangia of Female and Male Adults of the Ambrosia Beetle Platypus cylindrus Fab. (Coleoptera: Curculionidae). Insects 2021, 12, 881. https://doi.org/10.3390/insects12100881
Nones S, Simões F, Trindade CS, Matos J, Sousa E. Microbiome Associated with the Mycangia of Female and Male Adults of the Ambrosia Beetle Platypus cylindrus Fab. (Coleoptera: Curculionidae). Insects. 2021; 12(10):881. https://doi.org/10.3390/insects12100881
Chicago/Turabian StyleNones, Stefano, Fernanda Simões, Cândida Sofia Trindade, José Matos, and Edmundo Sousa. 2021. "Microbiome Associated with the Mycangia of Female and Male Adults of the Ambrosia Beetle Platypus cylindrus Fab. (Coleoptera: Curculionidae)" Insects 12, no. 10: 881. https://doi.org/10.3390/insects12100881
APA StyleNones, S., Simões, F., Trindade, C. S., Matos, J., & Sousa, E. (2021). Microbiome Associated with the Mycangia of Female and Male Adults of the Ambrosia Beetle Platypus cylindrus Fab. (Coleoptera: Curculionidae). Insects, 12(10), 881. https://doi.org/10.3390/insects12100881