Phylogenetic Complexity of Morphologically Identified Anopheles squamosus in Southern Zambia
Abstract
:Simple Summary
Abstract
1. Introduction
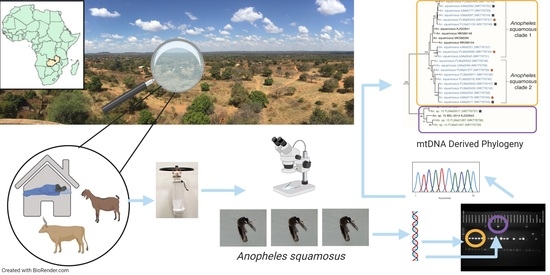
2. Materials and Methods
2.1. Study Area and Mosquito Collections
2.2. Sample Processing and Morphological Identification
2.3. Mosquito Species Assignment
2.4. Host Blood Meal Identification
2.5. Phylogenetic Analysis
3. Results
3.1. Species Composition
3.1.1. Collection Scheme I: Reactive-Test-and-Treat Program
3.1.2. Collection Scheme II: Transect
3.2. Host Identification
3.3. Species Assignment and Phylogenetic Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sturrock, H.J.W.; Roberts, K.W.; Wegbreit, J.; Ohrt, C.; Gosling, R.D. Tackling imported malaria: An elimination endgame. Am. J. Trop. Med. Hyg. 2015, 93, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Killeen, G.F. Characterizing, controlling and eliminating residual malaria transmission. Malar. J. 2014, 13, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moiroux, N.; Gomez, M.B.; Pennetier, C.; Elanga, E.; Djenontin, A.; Chandre, F.; Djegbe, I.; Guis, H.; Corbel, V. Changes in Anopheles funestus biting behavior following universal coverage of long-lasting insecticidal nets in Benin. J. Infect. Dis. 2012, 206, 1622–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moiroux, N.; Damien, G.B.; Egrot, M.; Djenontin, A.; Chandre, F.; Corbel, V.; Killeen, G.F.; Pennetier, C. Human exposure to early morning Anopheles funestus biting behavior and personal protection provided by long-lasting insecticidal nets. PLoS ONE 2014, 9, e104967. [Google Scholar] [CrossRef]
- Russell, T.L.; Govella, N.J.; Azizi, S.; Drakeley, C.J.; Kachur, S.P.; Killeen, G.F. Increased proportions of outdoor feeding among residual malaria vector populations following increased use of insecticide-treated nets in rural Tanzania. Malar. J. 2011, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Fornadel, C.M.; Norris, L.C.; Franco, V.; Norris, D.E. Unexpected anthropophily in the potential secondary malaria vectors Anopheles coustani s.l. and Anopheles squamosus in Macha, Zambia. Vector-Borne Zoonotic Dis. 2011, 11, 1173–1179. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, J.C.; Simubali, L.; Mbambara, S.; Musonda, M.; Mweetwa, S.; Mudenda, T.; Pringle, J.C.; Jones, C.M.; Norris, D.E. Detection of Plasmodium falciparum infection in Anopheles squamosus (Diptera: Culicidae) in an area targeted for malaria elimination, Southern Zambia. J. Med. Entomol. 2016, 53, 1482–1487. [Google Scholar] [CrossRef] [Green Version]
- Lacan, A. Anophelines of the Plateau Region of Madagascar in 1952. Mem. Sci. Madag. 1953, 4, 503–519. [Google Scholar]
- Subbarao, S.K.; Nanda, N.; Rahi, M.; Raghavendra, K. Biology and bionomics of malaria vectors in India: Existing information and what more needs to be known for strategizing elimination of malaria. Malar. J. 2019, 18, 396. [Google Scholar] [CrossRef]
- Moss, W.J.; Hamapumbu, H.; Kobayashi, T.; Shields, T.; Kamanga, A.; Clennon, J.; Mharakurwa, S.; Thuma, P.E.; Glass, G. Use of remote sensing to identify spatial risk factors for malaria in a region of declining transmission: A cross-sectional and longitudinal community survey. Malar. J. 2011, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mharakurwa, S.; Thuma, P.E.; Norris, D.E.; Mulenga, M.; Chalwe, V.; Chipeta, J.; Munyati, S.; Mutambu, S.; Mason, P.R. Malaria epidemiology and control in Southern Africa. Acta Trop. 2012, 121, 202–206. [Google Scholar] [CrossRef] [Green Version]
- Norris, L.C.; Norris, D.E. Heterogeneity and changes in inequality of malaria risk after introduction of insecticide-treated bed nets in Macha, Zambia. Am. J. Trop. Med. Hyg. 2013, 88, 710–717. [Google Scholar] [CrossRef] [Green Version]
- Kent, R.J.; Thuma, P.E.; Mharakurwa, S.; Norris, D.E. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am. J. Trop. Med. Hyg. 2007, 76, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tabue, R.N.; Awono-Ambene, P.; Etang, J.; Atangana, J.; Antonio-Nkondjio, C.; Toto, J.C.; Patchoke, S.; Leke, R.G.F.; Fondjo, E.; Mnzava, A.P.; et al. Role of Anopheles (Cellia) rufipes (Gough, 1910) and other local anophelines in human malaria transmission in the northern savannah of Cameroon: A cross-sectional survey. Parasites Vectors 2017, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillies, M.T. The role of secondary vectors of malaria in North-East Tanganyika. Trans. R Soc. Trop. Med. Hyg. 1964, 58, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Nepomichene, T.N.; Tata, E.; Boyer, S. Malaria case in Madagascar, probable implication of a new vector, Anopheles coustani. Malar. J. 2015, 14, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theobald, F.V. A Monograph of the Culicidae or Mosquitoes; British Museum of Natural History: London, UK, 1901. [Google Scholar]
- Wellman, F.C. Notes on the Common Mosquitoes of Bihe and Bailundo Districts, Portuguese West Africa. J. Infect. Dis. 1905, 2, 627–631. [Google Scholar] [CrossRef]
- Theobald, F.V. A Monograph of the Culicidae or Mosquitoes; British Museum of Natural History: London, UK, 1903. [Google Scholar]
- Kyalo, D.; Amratia, P.; Mundia, C.W.; Mbogo, C.M.; Coetzee, M.; Snow, R.W. A geo-coded inventory of anophelines in the Afrotropical Region south of the Sahara: 1898–2016. Wellcome Open Res. 2017, 2, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouznetsov, R.; World Health Organization. Distribution of Anophelines in the Yemen Arab Republic and Its Relation to Malaria; World Health Organization: Geneva, Switzerland, 1976. [Google Scholar]
- Barbier, D.; Moreau, J.P.; Radanielina, R. Epidemiologic study among companies of the armed-forces of Southeastern Madagascar Malagasy-Republic: Part III. Arch. Pasteur Madag. 1975, 44, 127–130. [Google Scholar]
- Nigatu, W.; Petros, B.; Lulu, M.; Adugna, N.; Wirtz, R. Species composition, feeding and resting behaviour of the common anthropophilic anopheline mosquitoes in relation to malaria transmission in Gambella, south west Ethiopia. Int. J. Trop. Insect Sci. 1994, 15, 371–377. [Google Scholar] [CrossRef]
- Degefa, T.; Zeynudin, A.; Godesso, A.; Michael, Y.H.; Eba, K.; Zemene, E.; Emana, D.; Birlie, B.; Tushune, K.; Yewhalaw, D. Malaria incidence and assessment of entomological indices among resettled communities in Ethiopia: A longitudinal study. Malar. J. 2015, 14, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansfield-Aders, W. Notes on malaria and filariasis in the Zanzibar Protectorate. Trans. R. Soc. Trop. Med. Hyg. 1927, 21, 207–214. [Google Scholar] [CrossRef]
- Jadin, J. Malaria control by residual DDT spraying in the Astrida Region, Ruanda-Urundl. In Annales de la Societe Belge de Medecine Tropicale; Societe Belge de Medecine Tropicale: Brussels, Belgium, 1951; pp. 631–651. [Google Scholar]
- Lumsden, W. Probable insect vectors of yellow fever virus, from monkey to man, in Bwamba County, Uganda. Bull. Entomol. Res. 1951, 42, 317–330. [Google Scholar] [CrossRef]
- Nepomichene, T.N.J.J.; Elissa, N.; Cardinale, E.; Boyer, S. Species diversity, abundance, and host preferences of mosquitoes (Diptera: Culicidae) in two different ecotypes of Madagascar with recent RVFV transmission. J. Med. Entomol. 2015, 52, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Hicks, E. The transmission of Wuchereria bancrofti in Sierra Leone. Ann. Trop. Med. Parasitol. 1932, 26, 407–422. [Google Scholar] [CrossRef]
- Monier, H. Faits nouveaux concernant l’épidémiologie du paludisme à Tananarive. Bulletin Société Pathol. Exot. 1935, 28, 775–778. [Google Scholar]
- Lewis, D.J. The egg of Anopheles squamosus Theobald. E. Afr. Med. J. 1946, 23, 18. [Google Scholar]
- Colbourne, M.; Wright, F. Malaria in the Gold Coast. W. Afr. Med. J. 1955, 4, 161–174. [Google Scholar]
- Diagne, N.; Fontenille, D.; Konate, L.; Faye, O.; Lamizana, M.T.; Legros, F.; Molez, J.F.; Trape, J.F. The Anopheles of Senegal-an annotated and illustrated check list. Bull. Soc. Pathol. Exot. 1994, 87, 267–277. [Google Scholar]
- Tantely, M.L.; Rakotoniaina, J.C.; Tata, E.; Andrianaivolambo, L.; Razafindrasata, F.; Fontenille, D.; Elissa, N. Biology of mosquitoes that are potential vectors of Rift Valley fever virus in different biotopes of the Central Highlands of Madagascar. J. Med Entomol. 2013, 50, 603–610. [Google Scholar] [CrossRef]
- Norris, L.C.; Norris, D.E. Phylogeny of anopheline (Diptera: Culicidae) species in southern Africa, based on nuclear and mitochondrial genes. J. Vector Ecol. 2015, 40, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Tantely, M.L.; Le Goff, G.; Boyer, S.; Fontenille, D. An updated checklist of mosquito species (Diptera: Culicidae) from Madagascar. Parasite 2016, 23. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M. Observations on nulliparous and parous rates in some common East African mosquitoes. Ann. Trop. Med. Parasitol. 1963, 57, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.T.; DeMeillon, B. The Anophelinae South of the Sahara (Ethiopian Zoogeographical Region); South African Institute for Medical Research: Johannesburg, South Africa, 1968.
- De Meillon, B. The Anophelini of the Ethiopian Geographical Region; Publications of the South African Institute for Medical Research: Johannesburg, South Africa, 1947; pp. 238–241. [Google Scholar]
- Lobo, N.F.; Laurent, B.S.; Sikaala, C.H.; Hamainza, B.; Chanda, J.; Chinula, D.; Krishnankutty, S.M.; Mueller, J.D.; Deason, N.A.; Hoang, Q.T.; et al. Unexpected diversity of Anopheles species in Eastern Zambia: Implications for evaluating vector behavior and interventions using molecular tools. Sci. Rep. 2015, 5, 17952. [Google Scholar] [CrossRef]
- St Laurent, B.; Cooke, M.; Krishnankutty, S.M.; Asih, P.; Mueller, J.D.; Kahindi, S.; Ayoma, E.; Oriango, R.M.; Thumloup, J.; Drakeley, C.; et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan Highlands. Am. J. Trop. Med. Hyg. 2016, 94, 327–335. [Google Scholar] [CrossRef]
- Pringle, J.C.; Tessema, S.; Wesolowski, A.; Chen, A.; Murphy, M.; Carpi, G.; Shields, T.M.; Hamapumbu, H.; Searle, K.M.; Kobayashi, T.; et al. Genetic evidence of focal Plasmodium falciparum transmission in a pre-elimination setting in Southern Province, Zambia. J. Infect. Dis. 2019, 219, 1254–1263. [Google Scholar] [CrossRef]
- Gillies, T.; Coetzee, M. A Supplement to the Anophelinae of Africa South of the Sahara: Afrotropical Region; South African Institute for Medical Research: Johannesburg, South Africa, 1987.
- Norris, D.E.; Shurtleff, A.C.; Toure, Y.T.; Lanzaro, G.C. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae s.s. (Diptera: Culicidae). J. Med Entomol. 2001, 38, 336–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, J.A.; Brogdon, W.G.; Collins, F.H. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993, 49, 520–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciubotariu, I.I.; Jones, C.M.; Kobayashi, T.; Bobanga, T.; Muleba, M.; Pringle, J.C.; Stevenson, J.C.; Carpi, G.; Norris, D.E. Genetic diversity of Anopheles coustani (Diptera: Culiciade) in malaria transmission foci in southern and central Africa. J. Med Entomol. 2020, 57, 1782–1792. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Christina, K.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Tedrow, R.E.; Ratovonjato, J.; Walker, E.D.; Ratsimbasoa, A.C.; Zimmerman, P.A. A novel assay for simultaneous assessment of mammalian host blood, mosquito species, and Plasmodium spp. in the medically important Anopheles mosquitoes of Madagascar. Am. J. Trop. Med. Hyg. 2019, 100, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Lips, M. Congo (Ex-Belgian) Anophelines. 7. Some Species of the Cellia and Neocellia Groups. Riv. Parassitol. Roma 1962, 23, 107–134. [Google Scholar]
- Raharimalala, F.; Andrianinarivomanana, T.; Rakotondrasoa, A.; Collard, J.; Boyer, S. Usefulness and accuracy of MALDI-TOF mass spectrometry as a supplementary tool to identify mosquito vector species and to invest in development of international database. Med. Vet. Entomol. 2017, 31, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Small, S.T.; Labbe, F.; Lobo, N.F.; Koekemoer, L.L.; Sikaala, C.H.; Neafsey, D.E.; Hahn, M.W.; Fontaine, M.C.; Besansky, N.J. Radiation with reticulation marks the origin of a major malaria vector. Proc. Natl. Acad. Sci. USA 2020, 117, 31583–31590. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for Malaria Vector Control; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]


| Species | Number Collected | Mean Number per Trap Night | % of Total Collection |
|---|---|---|---|
| Outdoors (35 trap nights) | |||
| An. squamosus | 311 | 8.9 | 41.3 |
| An. rufipes | 182 | 5.2 | 24.2 |
| An. quadriannulatus | 69 | 2.0 | 9.2 |
| An. arabiensis | 40 | 1.1 | 5.3 |
| An. coustani | 38 | 1.1 | 5.0 |
| An. longipalpis | 27 | 0.8 | 3.6 |
| An. pretoriensis | 1 | 0.03 | 0.1 |
| Unverified | 83 | 2.4 | 11.1 |
| Total | 751 | 21.5 | 100.0 |
| Indoors (61 trap nights) | |||
| An. arabiensis | 82 | 1.3 | 43.2 |
| An. squamosus | 54 | 0.9 | 28.4 |
| An. longipalpis | 7 | 0.1 | 3.7 |
| An. coustani | 6 | 0.1 | 3.2 |
| An. quadriannulatus | 4 | 0.07 | 2.1 |
| An. rufipes | 3 | 0.05 | 1.6 |
| Unverified | 34 | 0.6 | 17.9 |
| Total | 190 | 3.1 | 100.0 |
| Morphological Species | Number Collected | Mean Number per Trap Night | % of Total Collection |
|---|---|---|---|
| Outdoors (124 trap nights) | |||
| An. rufipes | 731 | 5.9 | 29.7 |
| An. coustani | 235 | 1.9 | 9.5 |
| An. squamosus | 194 | 1.6 | 7.9 |
| An. gambiae s.l. | 174 | 1.4 | 7.1 |
| An. funestus s.l. | 152 | 1.2 | 6.2 |
| An. pretoriensis | 92 | 0.7 | 3.7 |
| An. longipalpis | 60 | 0.5 | 2.4 |
| An. maculipalpis | 15 | 0.1 | 0.6 |
| An. theileri | 5 | 0.04 | 0.2 |
| An. brunnipes | 1 | 0.01 | 0.04 |
| An. dancalicus | 1 | 0.01 | 0.04 |
| An. hancocki/brohieri | 1 | 0.01 | 0.04 |
| An. machardyi | 1 | 0.01 | 0.04 |
| Unverified | 799 | 6.4 | 32.4 |
| Total | 2463 | 19.9 | 100.0 |
| Indoors (127 trap nights) | |||
| An. gambiae s.l. | 66 | 0.5 | 39.0 |
| An. rufipes | 23 | 0.2 | 13.6 |
| An. funestus s.l. | 7 | 0.1 | 4.1 |
| An. squamosus | 6 | 0.05 | 3.6 |
| An. coustani | 6 | 0.05 | 3.6 |
| An. longipalpis | 3 | 0.02 | 1.8 |
| Male | 1 | 0.01 | 0.6 |
| Unverified | 57 | 0.4 | 33.7 |
| Total | 169 | 1.3 | 100.0 |
| Human | Cow | Goat | |
|---|---|---|---|
| An. squamosus | 26 | 53 | |
| An. sp. 15 | 1 | 3 | |
| An. arabiensis | 3 | ||
| An. rufipes | 1 | 2 | |
| An. coustani | 2 | ||
| An. maculipalpis | 1 | ||
| An. quadriannulatus | 1 |
| Specimen ID | Molecular Species Identification | Morphological Species Identification | Accession Number | Month/Season of Collection | Collection Scheme/Trap Location | Host | An. Squamosus Clade |
|---|---|---|---|---|---|---|---|
| IcMa0238 | An. arabiensis | An. dancalicus | MK776730 | February/Rainy | I/Indoors | ||
| FLMa00483 | An. coustani | An. squamosus | MK776731 | May/Cool Dry | II/Goat Pen | Goat | |
| FLMa00475 | An. maculipalpis | An. squamosus | MK776733 | May/Cool Dry | II/Goat Pen | ||
| FLMa00485 | An. maculipalpis | An. squamosus | MK776734 | May/Cool Dry | II/Goat Pen | ||
| FLMa00431 | An. rufipes | An. squamosus | MK776735 | May/Cool Dry | II/Goat Pen | ||
| FLMa00456 | An. rufipes | An. squamosus | MK776736 | May/Cool Dry | II/Cattle Pen | Cow | |
| FLMa00017 | An. sp. 15 | An. squamosus | MK776737 | May/Cool Dry | II/Cattle Pen | Cow | |
| FLMa01287 | An. sp. 15 | An. squamosus | MK776738 | May/Cool Dry | II/Goat Pen | ||
| FLMa01407 | An. sp. 15 | An. squamosus | MK776739 | May/Cool Dry | II/Goat Pen | ||
| FLMa00433 | An. squamosus | An. squamosus | MK776741 | May/Cool Dry | II/Goat Pen | Goat | 1 |
| FLMa00465 | An. squamosus | An. squamosus | MK776743 | May/Cool Dry | II/Indoors | 1 | |
| FLMa00660 | An. squamosus | An. squamosus | MK776744 | May/Cool Dry | II/Goat Pen | Goat | 1 |
| FLMa01130 | An. squamosus | An. squamosus | MK776749 | May/Cool Dry | II/Goat Pen | Cow | 1 |
| IcMa0040 | An. squamosus | An. pretoriensis | MK776751 | February/Rainy | I/Cattle Pen | 1 | |
| IcMa0062 | An. squamosus | An. squamosus | MK776752 | February/Rainy | I/Cattle Pen | Cow | 1 |
| IcMa0097 | An. squamosus | An. squamosus | MK776754 | February/Rainy | I/Cattle Pen | Cow | 1 |
| IcMa0177 | An. squamosus | An. squamosus | MK776755 | February/Rainy | I/Goat Pen | 1 | |
| IcMa0221 | An. squamosus | An. squamosus | MK776757 | February/Rainy | I/Indoors | 1 | |
| FLMa00018 | An. squamosus | An. squamosus | MK776740 | May/Cool Dry | II/Cattle Pen | 2 | |
| FLMa00455 | An. squamosus | An. squamosus | MK776742 | May/Cool Dry | II/Cattle Pen | 2 | |
| FLMa00922 | An. squamosus | An. squamosus | MK776746 | May/Cool Dry | II/Goat Pen | 2 | |
| FLMa00932 | An. squamosus | An. squamosus | MK776747 | May/Cool Dry | II/Goat Pen | Cow | 2 |
| FLMa00971 | An. squamosus | An. squamosus | MK776748 | May/Cool Dry | II/Goat Pen | 2 | |
| FLMa01277 | An. squamosus | An. squamosus | MK776750 | May/Cool Dry | II/Goat Pen | Goat | 2 |
| IcMa0077 | An. squamosus | An. squamosus | MK776753 | February/Rainy | I/Cattle Pen | Cow | 2 |
| IcMa0179 | An. squamosus | An. squamosus | MK776756 | February/Rainy | I/Goat Pen | Goat | 2 |
| IcMa0232 | An. squamosus | An. squamosus | MK776758 | February/Rainy | I/Indoors | 2 | |
| IcMa0249 | An. squamosus | Male | MK776759 | February/Rainy | I/Indoors | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffman, J.E.; Ciubotariu, I.I.; Simubali, L.; Mudenda, T.; Moss, W.J.; Carpi, G.; Norris, D.E.; Stevenson, J.C.; on behalf of Southern and Central Africa International Centers of Excellence for Malaria Research. Phylogenetic Complexity of Morphologically Identified Anopheles squamosus in Southern Zambia. Insects 2021, 12, 146. https://doi.org/10.3390/insects12020146
Hoffman JE, Ciubotariu II, Simubali L, Mudenda T, Moss WJ, Carpi G, Norris DE, Stevenson JC, on behalf of Southern and Central Africa International Centers of Excellence for Malaria Research. Phylogenetic Complexity of Morphologically Identified Anopheles squamosus in Southern Zambia. Insects. 2021; 12(2):146. https://doi.org/10.3390/insects12020146
Chicago/Turabian StyleHoffman, Jordan E., Ilinca I. Ciubotariu, Limonty Simubali, Twig Mudenda, William J. Moss, Giovanna Carpi, Douglas E. Norris, Jennifer C. Stevenson, and on behalf of Southern and Central Africa International Centers of Excellence for Malaria Research. 2021. "Phylogenetic Complexity of Morphologically Identified Anopheles squamosus in Southern Zambia" Insects 12, no. 2: 146. https://doi.org/10.3390/insects12020146
APA StyleHoffman, J. E., Ciubotariu, I. I., Simubali, L., Mudenda, T., Moss, W. J., Carpi, G., Norris, D. E., Stevenson, J. C., & on behalf of Southern and Central Africa International Centers of Excellence for Malaria Research. (2021). Phylogenetic Complexity of Morphologically Identified Anopheles squamosus in Southern Zambia. Insects, 12(2), 146. https://doi.org/10.3390/insects12020146