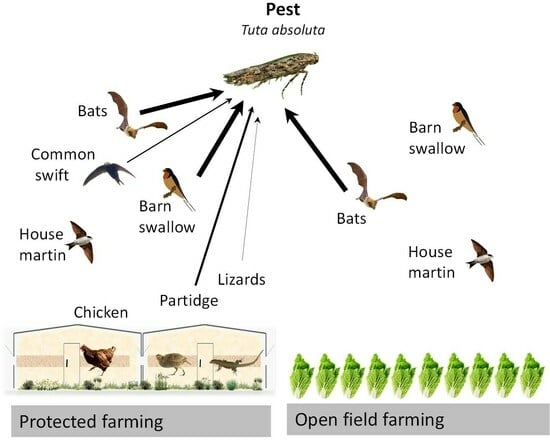
Tuta absoluta-Specific DNA in Domestic and Synanthropic Vertebrate Insectivore Feces
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects and Feces Collection
2.2. DNA Extraction, Quantification and Quality Control
2.3. Detection of Tuta absoluta in Domestic and Wild Animals
2.4. Real-Time PCR and Sensitivity Analysis
3. Results
3.1. Real-Time PCR and Sensitivity Analysis
3.2. DNA Yield and Efficiency of Tuta absoluta DNA Purification
3.3. Tuta absoluta DNA in Domestic and Wild Vertebrate Animals
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Organization of the United Nations. 2019. Available online: http://www.fao.org/faostat/en/#home (accessed on 1 February 2021).
- Gullino, M.L.; Albajes, R.; Nicot, P.C. Integrated Pest and Disease Management in Greenhouse Crops; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Jones, J.B.; Zitter, T.A.; Momol, M.T.; Miller, S.A. Compendium of Tomato Diseases and Pests; American Phytopathological Society: St. Paul, MN, USA, 2014; p. 168. [Google Scholar]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.G.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Catalan Ruescas, D.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest Sci. 2010, 3, 197–215. [Google Scholar] [CrossRef]
- Caparros Mejido, R.; Haubruge, E.; Verheggen, F.J. First evidence of deuterotokous parthenogenesis in the tomato leafminer, Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae). J. Pest Sci. 2012, 85, 409–412. [Google Scholar] [CrossRef]
- Biondi, A.; Guedes, R.N.C.; Wan, F.H.; Desneux, N. Ecology, worldwide spread, and management of the invasive South American tomato pinworm, Tuta absoluta: Past, present, and future. Annu. Rev. Entomol. 2018, 63, 239–258. [Google Scholar] [CrossRef] [PubMed]
- EPPO. Data sheets on quarantine pests: Tuta absoluta. EPPO Bull. 2005, 35, 434–435. [Google Scholar] [CrossRef]
- Guedes, R.N.C.; Roditakis, E.; Campos, M.R.; Haddi, K.; Bielza, P.; Siqueira, H.A.A.; Nauen, R. Insecticide resistance in the tomato pinworm Tuta absoluta: Patterns, spread, mechanisms, management and outlook. J. Pest Sci. 2019, 92, 1329–1342. [Google Scholar] [CrossRef]
- Desneux, N.; Han, P.; Mansour, R.; Arnó, J.; Brévault, T.; Campos, M.R.; Chailleux, A.; Guedes, R.N.; Karimi, J.; Konan, K.A.J.; et al. Integrated pest management of Tuta absoluta: Practical implementations across different world regions. J. Pest Sci. 2021, 95, 17–39. [Google Scholar] [CrossRef]
- Cherif, A.; Verheggen, F. A review of Tuta absoluta (Lepidoptera: Gelechiidae) host plants and their impact on management strategies. Biotechnol. Agron. Soc. Environ. 2019, 23, 270–278. [Google Scholar] [CrossRef]
- Tonnang, H.E.; Mohamed, S.F.; Khamis, F.; Ekesi, S. Identification and risk assessment for worldwide invasion and spread of Tuta absoluta with a focus on Sub-Saharan Africa: Implications for phytosanitary measures and management. PLoS ONE 2015, 10, e0135283. [Google Scholar] [CrossRef]
- Campos, M.R.; Biondi, A.; Adiga, A.; Guedes, R.N.; Desneux, N. From the Western Palaearctic region to beyond: Tuta absoluta 10 years after invading Europe. J. Pest Sci. 2017, 90, 787–796. [Google Scholar] [CrossRef]
- Guedes, N.C.; Picanco, M. Tuta absoluta in South America: Pest status, management & insecticide resistance. In Proceedings of the EPPO/IOBC/FAO/NEPPO Joint International Symposium on Management of Tuta absoluta (tomato borer), Agadir, Marocco, 16–18 November 2011; pp. 15–16. [Google Scholar]
- Cock, M.J.W.; van Lenteren, J.C.; Brodeur, J.; Barratt, B.I.P.; Bigler, F.; Bolckmans, K.; Cônsoli, F.L.; Haas, F.; Mason, P.G.; Parra, J.R.P. Do new access and benefit sharing procedures under the convention on biological diversity threaten the future of biological control? Biocontrol 2010, 55, 199–218. [Google Scholar] [CrossRef]
- Ferracini, C.; Bueno, V.H.P.; Dindo, M.L.; Ingegno, B.L.; Luna, M.G.; Salas Gervassio, N.G.; Sánchez, N.E.; Siscaro, G.; van Lenteren, J.C.; Zappalà, L.; et al. Natural enemies of Tuta absoluta in the Mediterranean basin, Europe and South America. Biocontrol Sci. Technol. 2019, 29, 578–609. [Google Scholar] [CrossRef]
- Cajamar. Análisis de la Campaña Hortofrutícola. Campaña 2020/2021, Fundación Cajamar/Caja Rural ed.; Cajamar Caja: Almería, Spain, 2021; Available online: https://publicacionescajamar.es/series-tematicas/informes-coyuntura-analisis-de-campana/analisis-de-la-campana-hortofruticola-de-almeria-campana-2020-2021/ (accessed on 27 July 2023).
- Schmidt, J.M.; Acebes-Doria, A.; Blaauw, B.; Kheirodin, A.; Pandey, S.; Lennon, K.; Grabarczyk, E.E. Identifying molecular-based trophic interactions as a resource for advanced integrated pest management. Insects 2021, 12, 358. [Google Scholar] [CrossRef]
- Díaz-Siefer, P.; Olmos-Moya, N.; Fontúrbel, F.E.; Lavandero, B.; Pozo, R.A.; Celis-Diez, J.L. Bird-mediated effects of pest control services on crop productivity: A global synthesis. J. Pest Sci. 2022, 95, 567–576. [Google Scholar] [CrossRef]
- Cohen, Y.; Bar-David, S.; Nielsen, M.; Bohmann, K.; Korine, C. An appetite for pests: Synanthropic insectivorous bats exploit cotton pest irruptions and consume various deleterious arthropods. Mol. Ecol. 2020, 29, 1185–1198. [Google Scholar] [CrossRef]
- European Commision. The Tomato Market in the EU: Vol. 1: Production, Areas and Yields; European Commission: Brussels, Belgium, 2019. [Google Scholar]
- Walsh, P.S.; Metzger, A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 1991, 10, 506–513. [Google Scholar] [CrossRef] [Green Version]
- Zink, F.A.; Tembrock, L.R.; Timm, A.E.; Gilligan, T.M. A Real-Time PCR Assay for Rapid Identification of Tuta absoluta (Lepidoptera: Gelechiidae). J. Econ. Entomol. 2020, 113, 1479–1485. [Google Scholar] [CrossRef] [Green Version]
- Barr, N.B.; Ledezma, L.A.; Farris, R.E.; Epstein, M.E.; Gilligan, T.M. A multiplex real-time polymerase chain reaction assay to diagnose Epiphyas postvittana (Lepidoptera: Tortricidae). J. Econ. Entomol. 2011, 104, 1706–1719. [Google Scholar] [CrossRef]
- Ledezma, L.A.; Barr, N.B.; Epstein, M.E.; Gilligan, T.M. Diagnosis of Lobesia botrana (Lepidoptera: Tortricidae) using real-time PCR. J. Econ. Entomol. 2016, 109, 1957–1962. [Google Scholar] [CrossRef]
- Gill, F.B. Ornithology, 3rd ed.; W.H. Freeman and Company: New York, NY, USA, 2007; pp. 1–12. [Google Scholar]
- Jedlicka, A. Vo ATE Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol. Ecol. Resour. 2014, 14, 1183–1197. [Google Scholar] [CrossRef]
- Regnaut, S.; Lucas, F.S.; Fumagalli, L. DNA degradation in avian faecal samples and feasibility of non-invasive genetic studies of threatened capercaillie populations. Conserv. Genet. 2006, 7, 449–453. [Google Scholar] [CrossRef] [Green Version]
- Zeale, M.R.K.; Butlin, R.; Barker, G.L.A.; Lees, D.C.L.; Jones, G. Taxon-Specific PCR for DNA Barcoding Arthropod Prey in Bat Faeces. Mol. Ecol. Resour. 2010, 11, 236–244. [Google Scholar] [CrossRef]
- Eriksson, P.; Mourkas, E.; González-Acuna, D.; Olsen, B.; Ellström, P. Evaluation and optimization of microbial DNA extraction from fecal samples of wild Antarctic bird species. Infect. Ecol. Epidemiol. 2017, 7, 1386536. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.; Cable, J.; Bruford, M.W. An evaluation of non-invasive sampling for genetic analysis in northern European reptiles. Herpetol. J. 2008, 18, 32–39. [Google Scholar]
- Oehm, J.; Juen, A.; Nagiller, K.; Neuhauser, S.; Traugott, M. Molecular scatology: How to improve prey DNA detection success in avian faeces? Mol. Ecol. Resour. 2011, 11, 620–628. [Google Scholar] [CrossRef]
- Cifuentes, D.; Chynoweth, R.; Bielza, P. Genetic study of Mediterranean and South American populations of tomato leafminer Tuta absoluta (Povolny, 1994) (Lepidoptera: Gelechiidae) using ribosomal and mitochondrial markers. Pest Manag. Sci. 2011, 67, 1155–1162. [Google Scholar] [CrossRef]
- Hódar, J.A.; Pleguezuelos, J.M.; Villafranca, C.; Fernández-Cardenete, J.R. Foraging mode of the Moorish gecko Tarentola mauritanica in an arid environment: Inferences from abiotic setting, prey availability and dietary composition. J. Arid. Environ. 2006, 65, 83–93. [Google Scholar] [CrossRef]
- Riccucci, M.; Lanza, B. Bats and insect pest control: A review. Vespertilio 2014, 17, 161–169. [Google Scholar]
- Rydell, J.; Entwistle, A.; Racey, P.A. Timing of foraging flights of three species of bats in relation to insect activity and predation risk. Oikos 1996, 76, 243–252. [Google Scholar] [CrossRef]
- McCracken, G.F.; Westbrook, J.K.; Brown, V.A.; Eldridge, M.; Federico, P.; Kunz, T.H. Bats track and exploit changes in insect pest populations. PLoS ONE 2012, 7, e43839. [Google Scholar] [CrossRef] [Green Version]
- de Torres, E.C.B.; Brown, V.A.; McCracken, G.F.; Kunz, T.H. Sympatric Bat Species Prey Opportunistically on a Major Moth Pest Pecans. Sustainability 2019, 11, 6365. [Google Scholar] [CrossRef] [Green Version]
- Weier, S.M.; Moodley, Y.; Fraser, M.F.; Linden, V.M.G.; Grass, I.; Tscharntke, T.; Taylor, P.J. Insect pest consumption by bats in macadamia orchards established by molecular diet analyses. Glob. Ecol. Conserv. 2019, 18, e00626. [Google Scholar] [CrossRef]
- Kemp, J.; Lopez-Baucells, A.; Rocha, R.; Wangensteen, O.S.; Andriatafika, Z.; Nair, A.; Cabeza, M. Bats as potential suppressors of multiple agricultural pests: A case study from Madagascar. Agric. Ecosyst. Environ. 2019, 269, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Aizpurua, O.; Budinski, I.; Georgiakakis, P.; Gopalakrishnan, S.; Ibanez, C.; Mata, V.; Rebelo, H.; Russo, D.; Szodoray-Paradi, F.; Zhelyazkova, V.; et al. Agriculture shapes the trophic niche of a bat preying on multiple pest arthropods across Europe: Evidence from DNA metabarcoding. Mol. Ecol. 2018, 27, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Baroja, U.; Garin, I.; Aihartza, J.; Arrizabalaga-Escudero, A.; Vallejo, N.; Aldasoro, M.; Goiti, U. Pest consumption in a vineyard system by the lesser horseshoe bat (Rhinolophus hipposideros). PLoS ONE 2019, 14, e0219265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-San Pedro, A.; Allendes, J.L.; Beltrán, C.A.; Chaperon, P.N.; Saldarriaga-Córdoba, M.M.; Silva, A.X.; Grez, A.A. Quantifying ecological and economic value of pest control services provided by bats in a vineyard landscape of central Chile. Agric. Ecosyst. Environ. 2020, 302, 107063. [Google Scholar] [CrossRef]
- Orłowski, G.; Karg, J. Diet breadth and overlap in three sympatric aerial insectivorous birds at the same location. Bird Study 2013, 60, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Bryant, D.M.; Turner, A.K. Central place foraging by swallows (Hirundinidae): The question of load size. Anim. Behav. 1982, 30, 845–856. [Google Scholar] [CrossRef]
- van Deventer, P. Leaf miner threatens tomato growing in Europe (Crop protection). Fruits Veg. Technol. 2009, 9, 10–12. Available online: https://edepot.wur.nl/189532 (accessed on 20 January 2023).
- Coelho, M.C.F.; França, F.H. Biology, larva chaetotaxy and description of the pupa and adult moth the tomato. Pesqui. Agropecuária Bras. 1987, 22, 129–135. [Google Scholar]
- Uchoa-Fernandes, M.A.; della Lucia, T.M.C.; Vilela, E.F. Mating, oviposition and pupation of Scrobipalpuloides absoluta (Meyr.) (Lepidoptera: Gelechiidae). An. Soc. Entomol. Bras. 1995, 24, 159–164. [Google Scholar] [CrossRef]
- Sabbahi, R.; Khalil, A. The effectiveness of pheromone traps in controlling the tomato leafminer, Tuta absoluta, in the United Arab Emirates. J. Plant Dis. Prot. 2022, 129, 367–374. Available online: https://hal.archives-ouvertes.fr/hal-01548714 (accessed on 27 July 2023). [CrossRef]
- Waugh, D.R. Predation Strategies in Aerial Feeding Birds. Unpubl. Ph.D. Thesis, University of Stirling, Stirling, UK, 1978. [Google Scholar]
- Seibert, A.M.; Koblitz, J.C.; Denzinger, A.; Schnitzler, H.U. Scanning behavior in echolocating common pipistrelle bats (Pipistrellus pipistrellus). PLoS ONE 2013, 8, e60752. [Google Scholar] [CrossRef] [PubMed]
- Lack, D.; Owen, D.F. The food of the swift. J. Anim. Ecol. 1955, 24, 120–136. [Google Scholar] [CrossRef]
- Orłowski, G.; Karg, J.; Karg, G. Functional invertebrate prey groups reflect dietary responses to phenology and farming activity and pest control services in three sympatric species of aerially foraging insectivorous birds. PLoS ONE 2014, 9, e114906. [Google Scholar] [CrossRef]
- Charbonnier, Y.; Barbaro, L.; Theillout, A.; Jactel, H. Numerical and functional responses of forest bats to a major insect pest in pine plantations. PLoS ONE 2014, 9, e109488. [Google Scholar] [CrossRef] [Green Version]


| Name | Description | Sequence | Tm (°C) | Source |
|---|---|---|---|---|
| Ta-ITS-191F | ITS (diagnostic forward primer) | 5′-GCGAGCCAAGTGATCCAC-3‘ | 64.7 | [20] |
| Ta-ITS-345R | ITS (diagnostic reverse primer) | 5′-AGGCGACGATGTGTACGA-3‘ | 63.1 | [20] |
| Ta-ITS-286P | ITS (diagnostic probe) | 5′-FAM-CTTAGAATACAT-CGTGCAGGCAGC-BHQ-1-3‘ | 67.6 | [20] 1 |
| RT-18S-F2 | 18S (control forward primer) | 5′-ACCGCCCTAGTTCTAACCGTAAA-3‘ | 65.4 | [21] |
| RT-18S-R2 | 18S (control reverse primer) | 5′-CCGCCGAGCCATTGTAGTAA-3‘ | 66.9 | [21] |
| RT-18S-P2 | 18S (control probe) | 5′-Cy5-TGTCATCTAGC-GATCCGCCGA-BHQ-3-3‘ | 72.1 | [21] 1 |
| Treatment | DNA Extraction Methods | ||
|---|---|---|---|
| PowerFecal DNA Isolation Kit | Indispin Pathogen Kit | Chelex | |
| Physical disruption | yes | no | yes |
| Proteinase K | no | yes | no |
| Boil (100 °C) | no | no | yes |
| Ethanol precipitation | yes | yes | no |
| Method | Origin | DNA ng/μL (mean ± SD) 1 | Cq 2 | ||
|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | FAM | Cy5 | ||
| PowerFecal | P. pipistrellus | 12.4 ± 0.7 | 17.9 ± 0.9 | 29.46 | 28.79 |
| P. perdix | 5.4 ± 0.4 | 17.6 ± 2.3 | 25.99 | 27.05 | |
| H. rustica | 2.5 ± 1.5 | 13.9 ± 4.6 | 28.37 | 29.33 | |
| P. vitticeps | 4.0 ± 3.7 | 14.1 ± 3.7 | 23.8 | 21.95 | |
| Indispin | P. pipistrellus | 21.9 ± 3.0 | 22.3 ± 2.1 | 21.47 | 22.17 |
| P. perdix | 60.5 ± 2.6 | 101.3 ± 1.6 | 15.57 | 15.98 | |
| H. rustica | 309.5 ± 22.1 | 800.5 ± 14.3 | 24.88 | 25.34 | |
| P. vitticeps | 61.0 ± 3.9 | 53.6 ± 18.0 | 20.26 | 18.56 | |
| Chelex | P. pipistrellus | 1160.4 ± 22.4 | 1358.7 ± 16.3 | 20.05 | 21.54 |
| P. perdix | 1404.4 ± 42.8 | 1471.6 ± 6.9 | 28.01 | 27.79 | |
| H. rustica | 1269.9 ± 43.8 | 1683.2 ± 46.3 | 29.08 | 27.79 | |
| P. vitticeps | 503.9 ± 16.2 | 395.1 ± 7.2 | 19.42 | 23.02 | |
| Class | Species | Location | Sample ID * | Total General DNA (μg/uL) | Specific T. absoluta DNA (Cq) | |
|---|---|---|---|---|---|---|
| Latin Name | Common Name | Inside/Outside Tomato Greenhouse | ||||
| Aves | Perdix perdix | Partidge | Inside | 1 | 91.0 | 33.92 |
| Gallus domesticus | Chicken | Inside | 2 | 26.4 | - ** | |
| Coturnix coturnix | Common quail | Inside | 3 | 126.9 | 35.25 | |
| Apus apus | Common swift | Outside | 4 | 508.6 | 34.91 | |
| id. | id. | id. | 5 | 832.0 | 34.70 | |
| id. | id. | id. | 6 | 450.4 | - | |
| Delichon urbicum | House martin | Outside | 7 | 774.1 | - | |
| id. | id. | id. | 8 | 726.9 | - | |
| id. | id. | id. | 9 | 642.9 | - | |
| id. | id. | id. | 10 | 501.3 | - | |
| id. | id. | id. | 11 | 747.7 | - | |
| Hirundo rustica | Swallow | Outside | 12 | 678.9 | 31.83 | |
| id. | id. | id. | 13 | 612.0 | 30.17 | |
| id. | id. | id. | 14 | 187.3 | - | |
| id. | id. | id. | 15 | 559.0 | - | |
| Reptilia | Podarcis hispanicus | Iberian wall lizard | Inside | 16 | 66.8 | 35.47 |
| Tarentola mauritanica | Common wall gecko | Inside | 17 | 187.9 | 34.44 | |
| Psammodromus algirus | Algerian sand racer | Inside | 18 | 411.3 | 35.03 | |
| Mammalia | Pipistrellus pipistrellus | Pipistrelle microbat | Outside | 19 | 17.4 | 31.56 |
| id. | id. | id. | 20 | 42.1 | 31.94 | |
| id. | id. | id. | 21 | 54.1 | 34.31 | |
| id. | id. | id. | 22 | 60.0 | 33.65 | |
| id. | id. | id. | 23 | 53.7 | 33.58 | |
| id. | id. | id. | 24 | 178.4 | 29.53 | |
| id. | id. | id. | 25 | 149.9 | 27.13 | |
| id. | id. | id. | 26 | 45.9 | 31.77 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janssen, D.; González-Miras, E.; Rodríguez, E. Tuta absoluta-Specific DNA in Domestic and Synanthropic Vertebrate Insectivore Feces. Insects 2023, 14, 673. https://doi.org/10.3390/insects14080673
Janssen D, González-Miras E, Rodríguez E. Tuta absoluta-Specific DNA in Domestic and Synanthropic Vertebrate Insectivore Feces. Insects. 2023; 14(8):673. https://doi.org/10.3390/insects14080673
Chicago/Turabian StyleJanssen, Dirk, Emilio González-Miras, and Estefanía Rodríguez. 2023. "Tuta absoluta-Specific DNA in Domestic and Synanthropic Vertebrate Insectivore Feces" Insects 14, no. 8: 673. https://doi.org/10.3390/insects14080673
APA StyleJanssen, D., González-Miras, E., & Rodríguez, E. (2023). Tuta absoluta-Specific DNA in Domestic and Synanthropic Vertebrate Insectivore Feces. Insects, 14(8), 673. https://doi.org/10.3390/insects14080673