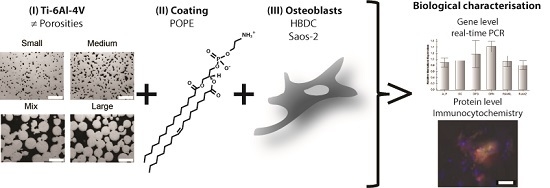
Comparison of the Influence of Phospholipid-Coated Porous Ti-6Al-4V Material on the Osteosarcoma Cell Line Saos-2 and Primary Human Bone Derived Cells
Abstract
:1. Introduction
2. Experimental Section
2.1. Sample Preparation
2.2. Phospholipid Coating
2.3. Cell Culture
2.4. Ribonucleic Acid (RNA) Extraction—Reverse Transcription Real-Time Polymerase Chain Reaction (RT-qPCR)
2.5. Statistical Analyses
2.6. Immunocytochemistry
3. Results
3.1. RT-qPCR
3.2. Immunocytochemistry
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Orthopedic devices market—global industry analysis, size, share, growth, trends and forecast, 2013–2019; ID: 2759691; Transparency Market Research: Albany, NY, USA, 2014.
- Wooley, P.H.; Schwarz, E.M. Aseptic loosening. Gene Ther. 2004, 11, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Okulov, I.V.; Pauly, S.; Kuhn, U.; Gargarella, P.; Marr, T.; Freudenberger, J.; Schultz, L.; Scharnweber, J.; Oertel, C.G.; Skrotzki, W.; et al. Effect of microstructure on the mechanical properties of as-cast Ti-Nb-Al-Cu-ni alloys for biomedical application. Mater. Sci. Eng. C Mater. Boil. Appl. 2013, 33, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Niinomi, M.; Akahori, T. Improvement of the fatigue life of titanium alloys for biomedical devices through microstructural control. Expert Rev. Med. Devices 2010, 7, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Surmenev, R.A.; Surmeneva, M.A.; Ivanova, A.A. Significance of calcium phosphate coatings for the enhancement of new bone osteogenesis—a review. Acta Biomater. 2014, 10, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Thorfve, A.; Lindahl, C.; Xia, W.; Igawa, K.; Lindahl, A.; Thomsen, P.; Palmquist, A.; Tengvall, P. Hydroxyapatite coating affects the Wnt signaling pathway during peri-implant healing in vivo. Acta Biomater. 2014, 10, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Pan, L.; Cui, F.; He, R.; Bao, H.; Wan, Q.; Wang, J. The recombinant human dentin matrix protein 1-coated titanium and its effect on the attachment, proliferation and ALP activity of MG63 cells. J. Mater. Sci. Mater. Med. 2012, 23, 2717–2726. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.W.; Lakshminarayanan, R.; Liu, S.P.; Goh, E.; Tan, D.; Beuerman, R.W.; Mehta, J.S. Dual functionalization of titanium with vascular endothelial growth factor and β-defensin analog for potential application in keratoprosthesis. J. Biomed. Mater. Res. Part B Appl. Biomater. 2012, 100B, 2090–2100. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.C.; Korn, P.; Stadlinger, B.; Range, U.; Möller, S.; Becher, J.; Schnabelrauch, M.; Mai, R.; Scharnweber, D.; Eckelt, U.; et al. Coating with artificial matrices from collagen and sulfated hyaluronan influences the osseointegration of dental implants. J. Mater. Sci. Mater. Med. 2013, 25, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lee, J.S.; Nemke, B.; Graf, B.K.; Royalty, K.; Illgen, R., 3rd; Vanderby, R., Jr.; Markel, M.D.; Murphy, W.L. Coating with a modular bone morphogenetic peptide promotes healing of a bone-implant gap in an ovine model. PLoS ONE 2012, 7, e50378. [Google Scholar] [CrossRef] [PubMed]
- Deing, A.; Luthringer, B.; Laipple, D.; Ebel, T.; Willumeit, R. A porous Ti-6Al-4V implant material for medical application. Int. J. Biomater. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Van der Stok, J.; Van der Jagt, O.P.; Amin Yavari, S.; De Haas, M.F.; Waarsing, J.H.; Jahr, H.; Van Lieshout, E.M.; Patka, P.; Verhaar, J.A.; Zadpoor, A.A.; et al. Selective laser melting-produced porous titanium scaffolds regenerate bone in critical size cortical bone defects. J. Orthop. Res. 2013, 31, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Espana, F.; Balla, V.K.; Bose, S.; Ohgami, Y.; Davies, N.M. Influence of porosity on mechanical properties and in vivo response of Ti6Al4V implants. Acta Biomater. 2010, 6, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Spoerke, E.D.; Murray, N.G.; Li, H.; Brinson, L.C.; Dunand, D.C.; Stupp, S.I. A bioactive titanium foam scaffold for bone repair. Acta Biomater. 2005, 1, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Staiger, M.P.; Pietak, A.M.; Huadmai, J.; Dias, G. Magnesium and its alloys as orthopedic biomaterials: A review. Biomaterials 2006, 27, 1728–1734. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Katz, J.L. Ultrasonic wave propagation in human cortical bone—П. Measurements of elastic properties and microhardness. J. Biomech. 1976, 9, 459–464. [Google Scholar] [CrossRef]
- Lee, T.; Lakes, R.S.; Lal, A. Investigation of bovine bone by resonant ultrasound spectroscopy and transmission ultrasound. Biomech. Model. Mechanobiol. 2002, 1, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.H.; Wang, T.; Burr, D.B. Shear strength and fatigue properties of human cortical bone determined from pure shear tests. Calcif. Tissue Int. 2001, 69, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Kemper, A.; McNally, C.; Kennedy, E.; Manoogian, S.; Duma, S. The material properties of human tibia cortical bone in tension and compression: Implications for the tibia index. In Proceedings of the 20th International Technical Conference on the Enhanced Safety of Vehicles Conference (ESV), Lyon, France, 2007.
- Matassi, F.; Botti, A.; Sirleo, L.; Carulli, C.; Innocenti, M. Porous metal for orthopedics implants. Clin. Cases Miner. Bone Metab. 2013, 10, 111–115. [Google Scholar] [PubMed]
- Alvarez, K.; Nakajima, H. Metallic scaffolds for bone regeneration. Materials 2009, 2, 790–832. [Google Scholar] [CrossRef]
- Ponader, S.; Von Wilmowsky, C.; Widenmayer, M.; Lutz, R.; Heinl, P.; Korner, C.; Singer, R.F.; Nkenke, E.; Neukam, F.W.; Schlegel, K.A. In vivo performance of selective electron beam-melted Ti-6Al-4V structures. J. Biomed. Mater. Res. A 2009, 92, 56–62. [Google Scholar]
- Li, J.P.; Habibovic, P.; van den Doel, M.; Wilson, C.E.; de Wijn, J.R.; Van Blitterswijk, C.A.; De Groot, K. Bone ingrowth in porous titanium implants produced by 3D fiber deposition. Biomaterials 2007, 28, 2810–2820. [Google Scholar] [CrossRef] [PubMed]
- Faria, P.E.; Carvalho, A.L.; Felipucci, D.N.; Wen, C.; Sennerby, L.; Salata, L.A. Bone formation following implantation of titanium sponge rods into humeral osteotomies in dogs: A histological and histometrical study. Clin. Implant. Dent. Relat. Res. 2008, 12, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Jorge, H. Compounding and processing of a water soluble binder for powder injection moulding. Ph.D. Thesis, Universidade do Minho, Braga, Portugal, 2008. [Google Scholar]
- Willumeit, R.; Schuster, A.; Iliev, P.; Linser, S.; Feyerabend, F. Phospholipids as implant coatings. J. Mater. Sci. Mater. Med. 2007, 18, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Luthringer, B.J.; Katha, U.M.; Willumeit, R. Phosphatidylethanolamine biomimetic coating increases mesenchymal stem cell osteoblastogenesis. J. Mater. Sci. Mater. Med. 2014, 25, 2561–2571. [Google Scholar] [CrossRef] [PubMed]
- Zachowski, A. Phospholipids in animal eukaryotic membranes: Transverse asymmetry and movement. Biochem. J. 1993, 294, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kung, C. A possible unifying principle for mechanosensation. Nature 2005, 436, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Sprong, H.; Van der Sluijs, P.; Van Meer, G. How proteins move lipids and lipids move proteins. Nature 2001, 2, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Wuthier, R.E. Lipid composition of isolated epiphyseal cartilage cells, membranes and matrix vesicles. Biochim. Biophys. Acta 1975, 409, 128–143. [Google Scholar] [CrossRef]
- Camolezi, F.L.; Daghastanli, K.R.; Magalhaes, P.P.; Pizauro, J.M.; Ciancaglini, P. Construction of an alkaline phosphatase-liposome system: A tool for biomineralization study. Int. J. Biochem. Cell Boil. 2002, 34, 1091–1101. [Google Scholar] [CrossRef]
- Eanes, E.D. Mixed phospholipid liposome calcification. Bone Miner. 1992, 17, 269–272. [Google Scholar] [CrossRef]
- Letellier, S.R.; Lochhead, M.J.; Campbell, A.A.; Vogel, V. Oriented growth of calcium oxalate monohydrate crystals beneath phospholipid monolayers. Biochim. Biophys. Acta 1998, 1380, 31–45. [Google Scholar] [CrossRef]
- Santin, M. Calcium-binding phospolipids as a coating material for implant osteointergration. Interface 2006, 3, 277–281. [Google Scholar] [PubMed]
- Ishihara, K.; Nakabayashi, N.; Fukumoto, K.; Aoki, J. Improvement of blood compatibility on cellulose dialysis membrane. I. Grafting of 2-methacryloyloxyethyl phosphorylcholine on to a cellulose membrane surface. Biomaterials 1992, 13, 145–149. [Google Scholar] [CrossRef]
- Krishna, O.D.; Kim, K.; Byun, Y. Covalently grafted phospholipid monolayer on silicone catheter surface for reduction in platelet adhesion. Biomaterials 2005, 26, 7115–7123. [Google Scholar] [CrossRef] [PubMed]
- Emoto, K.; Inadome, H.; Kanaho, Y.; Narumiya, S.; Umeda, M. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Boil. Chem. 2005, 280, 37901–37907. [Google Scholar] [CrossRef] [PubMed]
- Willumeit, R.; Schossig, M.; Clemens, H.; Feyerabend, F. In vitro interactions of human chondrocytes and mesenchymal stem cells, and of mouse macrophages with phospholipid-covered metallic implant materials. Eur. Cell Mater. 2007, 13, 11–25. [Google Scholar] [PubMed]
- Golub, M.; Lott, D.; Watkins, E.; Garamus, V.; Luthringer, B.; Stoermer, M.; Schreyer, A.; Willumeit, R. X-ray and neutron investigation of self-assembled lipid layers on a titanium surface. Biointerphases 2013, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.A. Human osteoblast culture. Methods Mol. Med. 2003, 80, 3–18. [Google Scholar] [PubMed]
- Gartland, A.; Rumney, R.M.; Dillon, J.P.; Gallagher, J.A. Isolation and culture of human osteoblasts. Methods Mol. Biol. 2012, 806, 337–355. [Google Scholar] [PubMed]
- Burmester, A.; Luthringer, B.; Willumeit, R.; Feyerabend, F. Comparison of the reaction of bone-derived cells to enhanced MgCl2-salt concentrations. Biomatter 2014, 4, e967616. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. Qbase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s CT difference” formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Crockett, J.C.; Rogers, M.J.; Coxon, F.P.; Hocking, L.J.; Helfrich, M.H. Bone remodelling at a glance. J. Cell Sci. 2011, 124, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, R.T.; Ge, C.; Xiao, G.; Roca, H.; Jiang, D. Transcriptional regulation of osteoblasts. Cells Tissues Organs 2009, 189, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Niyibizi, C.; Eyre, D.R. Bone type V collagen: Chain composition and location of a trypsin cleavage site. Connect. Tissue Res. 1989, 20, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Rho, J.Y.; Kuhn-Spearing, L.; Zioupos, P. Mechanical properties and the hierarchical structure of bone. Med. Eng. Phys. 1998, 20, 92–102. [Google Scholar] [CrossRef]
- Marom, R.; Shur, I.; Solomon, R.; Benayahu, D. Characterization of adhesion and differentiation markers of osteogenic marrow stromal cells. J. Cell. Physiol. 2005, 202, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Z.; Yoshida, C.A.; Furuichi, T.; Amizuka, N.; Ito, M.; Fukuyama, R.; Miyazaki, T.; Kitaura, H.; Nakamura, K.; Fujita, T.; et al. RUNX2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 2007, 236, 1876–1890. [Google Scholar] [CrossRef] [PubMed]
- Aubin, J.E. Regulation of osteoblast formation and function. Rev. Endocr. Metab. Disord. 2001, 2, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Malaval, L.; Aubin, J.E. Global amplification polymerase chain reaction reveals novel transitional stages during osteoprogenitor differentiation. J. Cell Sci. 2003, 116, 1787–1796. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.S.; King, K.A.; Rabaia, N.A.; Pathare, P.; Srinivasan, S. Upregulation of osteopontin by osteocytes deprived of mechanical loading or oxygen. J. Bone Miner. Res. 2005, 20, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.L. Chapter 9—alkaline phosphatases. In Dynamics of bone and cartilage metabolism, 2nd ed.; Seibel, M.J., Robins, S.P., Bilezikian, J.P., Eds.; Academic Press: Burlington, MA, USA, 2006; pp. 153–164. [Google Scholar]
- Kitazawa, R.; Mori, K.; Yamaguchi, A.; Kondo, T.; Kitazawa, S. Modulation of mouse RANKL gene expression by Runx2 and vitamin D3. J. Cell. Biochem. 2008, 105, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Kawana, F.; Sasaki, T. Osteoclast differentiation and characteristic trabecular bone formation during growth plate destruction in osteoprotegerin-deficient mice. J. Electron. Microsc. 2003, 52, 515–525. [Google Scholar] [CrossRef]
- Lidington, E.A.; Moyes, D.L.; McCormack, A.M.; Rose, M.L. A comparison of primary endothelial cells and endothelial cell lines for studies of immune interactions. Transpl. Immunol. 1999, 7, 239–246. [Google Scholar] [CrossRef]
- Pautke, C.; Schieker, M.; Tischer, T.; Kolk, A.; Neth, P.; Mutschler, W.; Milz, S. Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 Os in comparison to human osteoblasts. Anticancer Res. 2004, 24, 3743–3748. [Google Scholar] [PubMed]
- Wang, X.F.; Zhang, Y.K.; Yu, Z.S.; Zhou, J.L. The role of the serum RANKL/OPG ratio in the healing of intertrochanteric fractures in elderly patients. Mol. Med. Reports 2013, 7, 1169–1172. [Google Scholar]
- Van Tuyl, L.H.; Voskuyl, A.E.; Boers, M.; Geusens, P.; Landewe, R.B.; Dijkmans, B.A.; Lems, W.F. Baseline RANKL:OPG ratio and markers of bone and cartilage degradation predict annual radiological progression over 11 years in rheumatoid arthritis. Ann. Rheum. Dis. 2010, 69, 1623–1628. [Google Scholar] [CrossRef] [PubMed]




| Material | Porosity | Mechanical Properties | O2 Content | ||||
|---|---|---|---|---|---|---|---|
| Porosity 2D | Porosity 3D | E | UTS (UCS) | YS (CYS) | εf (εc) | (µg/g) | |
| (%) | (%) | (GPa) | (MPa) | (MPa) | (%) | ||
| Small | 5 ± 1 | – | 101 ± 5 | 806 ± 2 | 707 ± 4 | 14 ± 2 | 1509 |
| – | – | 109 ± 3 | 1358 ± 37 | 783 ± 16 | 26 ± 1 | ||
| Medium | 11 ± 1 | – | 93 ± 5 | 733 ± 24 | 628 ± 13 | 5 ± 3 | 2013 |
| – | – | 92 ± 3 | 1341 ± 66 | 741 ± 18 | 26 ± 2 | ||
| Mixed | 33 ± 5 | 29 ± 1 | 31 ± 6 | 95 ± 40 | – | – | 1918 |
| – | – | 42 ± 2 | 623 ± 20 | 261 ± 10 | 25 ± 1 | ||
| Large | 34 ± 1 | 34 ± 1 | 18 ± 1 | 98 ± 10 | – | – | 1918 |
| – | – | 21 ± 2 | 306 ± 7 | 152 ± 5 | 14 ± 1 | ||
| Ti-6Al-4V ELI grade | – | – | 112 ± 7 | 860 | 795 | 10 | – |
| Natural bone | – | – | 10–30 a | 133 c | – | – | – |
| 8–33 b | 200 ± 20 d | 130–180 e | |||||
| Group | Target Name/NCBI RefSeq | |||
|---|---|---|---|---|
| Abbreviation | Forward | Reverse | Amplicon Length (bp) | |
| reference genes | actin, beta/NM_001101 | |||
| ACTB | CTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGT | 134 bp | |
| Ribosomal protein L10/NM_006013 | ||||
| RPL10 | AGTGGATGAGTTTCCGCTTT | ATATGGAAGCCATCTTTGCC | 135 bp | |
| target genes | Osteocalcin (or BGLAP, bone gamma-carboxyglutamate (gla) protein)/NM_199173 | |||
| OC | GGCAGCGAGGTAGTGAAGAG | CTGGAGAGGAGCAGAACTGG | 95 bp | |
| Osteoprotegerin (or TNFRSF11B, Tumour necrosis factor receptor superfamily member 11B)/NM_002546 | ||||
| OPG | CGCTCGTGTTTCTGGACAT | GGACATTTGTCACACAACAGC | 112 bp | |
| Osteopontin (or SPP1, secreted phosphoprotein 1)/NM_000582 | ||||
| OPN | CTCCATTGACTCGAACGACTC | CAGGTCTGCGAAACTTCTTAGAT | 230 bp | |
| Alkaline phosphatase, liver/bone/kidney/NM_000478 | ||||
| ALPL | CACCCACGTCGATTGCATCT | TAGCCACGTTGGTGTTGAGC | 211 bp | |
| Runt-related transcription factor 2/NM_001024630 | ||||
| RUNX2 | CAGTAGATGGACCTCGGGAA | ATACTGGGATGAGGAATGCG | 112 bp | |
| Control | Material | Gene | 3 W | 5 W | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Regulation | p-Value | Regulation | p-Value | |||||||
| Mirror polished | Small | ALP | −1.84 | ↔ | 4.96 × 10−2 | * | −1.13 | ↔ | 2.98 × 10−1 | |
| OC | −1.15 | ↔ | 3.97 × 10−1 | 1.28 | ↔ | 2.49 × 10−1 | ||||
| OPG | −1.6 | ↔ | 8.84 × 10−3 | ** | −1.21 | ↔ | 1.97 × 10−1 | |||
| OPN | −1.88 | ↔ | 5.72 × 10−3 | ** | −3.24 | ↓ | 2.72 × 10−1 | * | ||
| RANKL | −13.85 | ↓ | 2.44 × 10−2 | * | −1.16 | ↔ | 2.89 × 10−1 | |||
| RUNX2 | −1.29 | ↔ | 1.26 × 10−1 | 1.14 | ↔ | 1.75 × 10−1 | ||||
| Medium | ALP | −3.07 | ↓ | 2.89 × 10−3 | −1.33 | ↔ | 3.37 × 10−1 | |||
| OC | −1.29 | ↔ | 3.62 × 10−1 | ** | 1.62 | ↔ | 7.00 × 10−2 | |||
| OPG | −1.63 | ↔ | 2.84 × 10−2 | −1.24 | ↔ | 8.33 × 10−2 | ||||
| OPN | −1.88 | ↔ | 8.86 × 10−3 | * | −3.84 | ↓ | 2.27 × 10−2 | |||
| RANKL | −3.48 | ↓ | 1.24 × 10−1 | −1.33 | ↔ | 1.71 × 10−1 | * | |||
| RUNX2 | −1.48 | ↔ | 1.87 × 10−1 | 1.03 | ↔ | 7.72 × 10−1 | ||||
| Mix | ALP | −1.79 | ↔ | 2.14 × 10−2 | −1.55 | ↔ | 3.24 × 10−2 | |||
| OC | 1.04 | ↔ | 7.59 × 10−1 | * | 1.39 | ↔ | 9.49 × 10−2 | * | ||
| OPG | 1.02 | ↔ | 8.59 × 10−1 | 1.07 | ↔ | 5.10 × 10−2 | ||||
| OPN | −1.89 | ↔ | 1.52 × 10−3 | −8.61 | ↓ | 1.41 × 10−2 | ||||
| RANKL | −9.07 | ↓ | 3.01 × 10−3 | ** | −1.26 | ↔ | 4.94 × 10−1 | * | ||
| RUNX2 | 1.17 | ↔ | 5.96 × 10−1 | ** | 1.08 | ↔ | 5.58 × 10−1 | |||
| Large | ALP | −1.7 | ↔ | 6.69 × 10−2 | −1.02 | ↔ | 6.78 × 10−1 | |||
| OC | −1.11 | ↔ | 2.46 × 10−1 | 1.72 | ↔ | 5.38 × 10−2 | ||||
| OPG | −1.28 | ↔ | 5.31 × 10−2 | 1.08 | ↔ | 3.58 × 10−1 | ||||
| OPN | −1.91 | ↔ | 4.94 × 10−3 | −3.25 | ↓ | 4.10 × 10−2 | ||||
| RANKL | −2.92 | ↓ | 5.40 × 10−2 | ** | −1.18 | ↔ | 7.87 × 10−1 | * | ||
| RUNX2 | 1.4 | ↔ | 1.97 × 10−1 | 1.05 | ↔ | 5.32 × 10−1 | ||||
| Control | Material | Gene | 3 W | 5 W | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Regulation | p-Value | Regulation | p-Value | |||||||
| Mirror polished | Small | ALP | 2.35 | ↑ | 3.00 × 10−6 | *** | 1.57 | ↔ | 1.64 × 10−3 | ** |
| OC | 2.52 | ↑ | 3.00 × 10−6 | *** | 3.85 | ↑ | 1.26 × 10−2 | * | ||
| OPG | 3.95 | ↑ | 0.00 | *** | −1.38 | ↔ | 2.86 × 10−1 | |||
| OPN | 1.31 | ↔ | 4.98 × 10−1 | 1.06 | ↔ | 9.90 × 10−1 | ||||
| RANKL | 1.97 | ↔ | 2.08 × 10−3 | ** | 1.06 | ↔ | 9.39 × 10−1 | |||
| RUNX2 | 2.65 | ↑ | 1.05 × 10−4 | *** | 1.59 | ↔ | 1.45 × 10−1 | |||
| Medium | ALP | 1.61 | ↔ | 1.06 × 10−2 | * | 1.78 | ↔ | 1.41 × 10−3 | ** | |
| OC | 1.54 | ↔ | 3.93 × 10−2 | * | 2.56 | ↑ | 1.10 × 10−5 | *** | ||
| OPG | 1.75 | ↔ | 3.42 × 10−3 | ** | −1.2 | ↔ | 3.74 × 10−1 | |||
| OPN | 1.48 | ↔ | 1.08 × 10−1 | 1.14 | ↔ | 6.59 × 10−1 | ||||
| RANKL | 1.9 | ↔ | 1.91 × 10−2 | * | 2.76 | ↑ | 6.15 × 10−3 | ** | ||
| RUNX2 | 1.31 | ↔ | 7.85 × 10−2 | 1.79 | ↔ | 3.52 × 10−2 | * | |||
| Mix | ALP | 1.04 | ↔ | 7.23 × 10−1 | 1.23 | ↔ | 1.80 × 10−1 | |||
| OC | −3.79 | ↓ | 3.30 × 10−5 | *** | 1.34 | ↔ | 1.35 × 10−3 | ** | ||
| OPG | −1.3 | ↔ | 4.94 × 10−2 | * | −1.59 | ↔ | 1.82 × 10−2 | * | ||
| OPN | 2.55 | ↑ | 6.20 × 10−3 | ** | −1.13 | ↔ | 3.70 × 10−1 | |||
| RANKL | 1.22 | ↔ | 3.01 × 10−1 | 2.17 | ↑ | 1.90 × 10−2 | * | |||
| RUNX2 | 1.18 | ↔ | 2.36 × 10−1 | 1.54 | ↔ | 1.27 × 10−1 | ||||
| Large | ALP | −1.86 | ↔ | 1.20 × 10−4 | *** | 1.2 | ↔ | 2.15 × 10−1 | ||
| OC | −10.16 | ↓ | 1.00 × 10−6 | *** | 1.55 | ↔ | 6.90 × 10−5 | *** | ||
| OPG | −1.07 | ↔ | 5.82 × 10−1 | −1.37 | ↔ | 3.63 × 10−1 | ||||
| OPN | 2.2 | ↑ | 3.65 × 10−3 | ** | 1.13 | ↔ | 6.07 × 10−1 | |||
| RANKL | −1.53 | ↔ | 3.15 × 10−2 | * | 1.98 | ↔ | 7.05 × 10−2 | |||
| RUNX2 | 1.23 | ↔ | 6.40 × 10−2 | 1.79 | ↔ | 1.15 × 10−2 | * | |||
| Material | Gene | 3W | 5W | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Regulation | p-Value | Regulation | p-Value | ||||||
| Mirror Polished | ALP | 2.75 | ↑ | 5.87 × 10−4 | *** | 4.13 | ↑ | 1.30 × 10−3 | |
| OC | −1.45 | ↔ | 5.38 × 10−2 | 4.54 | ↑ | 2.00 × 10−6 | *** | ||
| OPG | −1.89 | ↔ | 4.07 × 10−1 | 1.61 | ↔ | 7.85 × 10−3 | ** | ||
| OPN | −2.13 | ↓ | 2.13 × 10−3 | ** | 33.62 | ↑ | 0.00 | ||
| RANKL | 2.02 | ↑ | 3.17 × 10−2 | * | 2.07 | ↑ | 3.30 × 10−3 | ** | |
| RUNX2 | 1.39 | ↔ | 2.30 × 10−1 | 2.13 | ↑ | 4.00 × 10−6 | *** | ||
| Small | ALP | 6.80 | ↑ | 1.27 × 10−4 | *** | 4.35 | ↑ | 2.20 × 10−5 | *** |
| OC | −1.61 | ↔ | 7.76 × 10−2 | 5.7 | ↑ | 0.00 | *** | ||
| OPG | −1.67 | ↔ | 1.77 × 10−2 | * | 1.74 | ↔ | 2.02 × 10−3 | ** | |
| OPN | −1.71 | ↔ | 1.64 × 10−3 | ** | 89.84 | ↑ | 0.00 | *** | |
| RANKL | 33.30 | ↑ | 1.30 × 10−5 | *** | 1.81 | ↔ | 2.27 × 10−3 | * | |
| RUNX2 | 1.71 | ↔ | 7.56 × 10−4 | *** | 1.46 | ↔ | 1.76 × 10−4 | *** | |
| Medium | ALP | 6.89 | ↑ | 1.00 × 10−6 | *** | 3.91 | ↑ | 0.00 | *** |
| OC | −1.60 | ↔ | 6.60 × 10−2 | 5.67 | ↑ | 8.00 × 10−6 | *** | ||
| OPG | −1.85 | ↔ | 1.10 × 10−1 | 2.85 | ↑ | 2.33 × 10−4 | *** | ||
| OPN | −1.72 | ↔ | 1.33 × 10−2 | * | 119.72 | ↑ | 0.00 | *** | |
| RANKL | 5.91 | ↑ | 6.40 × 10−3 | ** | 2 | ↔ | 7.67 × 10−3 | ** | |
| RUNX2 | 2.31 | ↑ | 2.81 × 10−3 | ** | 1.56 | ↔ | 1.85 × 10−2 | * | |
| Mix | ALP | 6.44 | ↑ | 0.00 | *** | 10.34 | ↑ | 1.00 × 10−6 | *** |
| OC | −1.23 | ↔ | 4.10 × 10−1 | 13.73 | ↑ | 8.73 × 10−3 | ** | ||
| OPG | −2.13 | ↓ | 4.48 × 10−3 | ** | 2.32 | ↑ | 1.22 × 10−3 | ** | |
| OPN | −2.04 | ↓ | 0.00 | *** | 392.33 | ↑ | 0.00 | *** | |
| RANKL | 22.95 | ↑ | 1.20 × 10−3 | ** | 3.36 | ↑ | 7.30 × 10−5 | *** | |
| RUNX2 | 1.72 | ↔ | 7.88 × 10−4 | *** | 2.56 | ↑ | 8.50 × 10−5 | *** | |
| Large | ALP | 5.35 | ↑ | 4.00 × 10−6 | *** | 3.19 | ↑ | 6.42 × 10−3 | ** |
| OC | −1.26 | ↔ | 4.29 × 10−1 | 10.53 | ↑ | 0.00 | *** | ||
| OPG | −1.50 | ↔ | 7.52 × 10−3 | ** | 2.54 | ↑ | 1.22 × 10−2 | * | |
| OPN | −1.65 | ↔ | 7.39 × 10−4 | *** | 145.41 | ↑ | 0.00 | *** | |
| RANKL | 6.18 | ↑ | 1.60 × 10−5 | *** | 2.9 | ↑ | 1.77 × 10−2 | * | |
| RUNX2 | 1.04 | ↔ | 8.19 × 10−1 | 2.27 | ↑ | 1.05 × 10−4 | *** | ||
| Material | Gene | 3 W | 5 W | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Regulation | p-Value | Regulation | p-Value | ||||||
| Mirror Polished | ALP | 1.82 | ↔ | 1.72 × 10−4 | *** | 3.89 | ↑ | 0.00 | *** |
| OC | −2.73 | ↓ | 1.87 × 10−4 | *** | 1.59 | ↔ | 1.13 × 10−2 | * | |
| OPG | −1.13 | ↔ | 2.68 × 10−1 | 3.49 | ↑ | 1.79 × 10−4 | *** | ||
| OPN | 1.26 | ↔ | 5.40 × 10−1 | 1.28 | ↔ | 3.27 × 10−1 | |||
| RANKL | 1.06 | ↔ | 9.08 × 10−1 | 4.58 | ↑ | 4.84 × 10−3 | ** | ||
| RUNX2 | 1.22 | ↔ | 1.36 × 10−1 | 1.56 | ↔ | 1.41 × 10−1 | |||
| Small | ALP | 1.17 | ↔ | 2.2 × 10−1 | 3.13 | ↑ | 1.80 × 10−5 | *** | |
| OC | −1.13 | ↔ | 3.87 × 10−1 | 1.15 | ↔ | 7.96 × 10−1 | |||
| OPG | −3.11 | ↓ | 0.00 | *** | 4.85 | ↑ | 3.99 × 10−3 | ** | |
| OPN | −1.48 | ↔ | 3.62 × 10−1 | 2.66 | ↑ | 3.77 × 10−4 | *** | ||
| RANKL | −2.54 | ↓ | 8.35 × 10−3 | 7.24 | ↑ | 9.00 × 10−6 | *** | ||
| RUNX2 | −1.75 | ↔ | 2.82 × 10−2 | −1.06 | ↔ | 4.39 × 10−1 | |||
| Medium | ALP | 1.31 | ↔ | 3.12 × 10−1 | 1.57 | ↔ | 3.14 × 10−3 | ** | |
| OC | −2.32 | ↓ | 1.99 × 10−3 | ** | 1.19 | ↔ | 2.14 × 10−1 | ||
| OPG | −1.49 | ↔ | 3.06 × 10−3 | ** | 2.56 | ↑ | 1.31 × 10−4 | *** | |
| OPN | −2.12 | ↓ | 2.49 × 10−2 | * | −1.16 | ↔ | 5.21 × 10−1 | ||
| RANKL | −1.86 | ↔ | 5.32 × 10−2 | 2.17 | ↑ | 2.09 × 10−3 | *** | ||
| RUNX2 | 1.35 | ↔ | 4.09 × 10−1 | 1 | ↔ | 9.63 × 10−1 | |||
| Mix | ALP | 1.29 | ↔ | 8.11 × 10−2 | 1.78 | ↔ | 2.64 × 10−3 | ** | |
| OC | −1.70 | ↔ | 5.91 × 10−2 | 1.51 | ↔ | 3.27 × 10−2 | * | ||
| OPG | 1.20 | ↔ | 6.56 × 10−2 | 3.51 | ↑ | 3.00 × 10−5 | *** | ||
| OPN | −1.55 | ↔ | 1.87 × 10−1 | −1.24 | ↔ | 5.28 × 10−2 | |||
| RANKL | −1.30 | ↔ | 1.21 × 10−1 | −1.09 | ↔ | 9.06 × 10−1 | |||
| RUNX2 | −1.13 | ↔ | 4.17 × 10−1 | 1.19 | ↔ | 5.26 × 10−1 | |||
| Large | ALP | 1.54 | ↔ | 4.96 × 10−2 | * | 1.68 | ↔ | 8.91 × 10−3 | ** |
| OC | −1.70 | ↔ | 6.18 × 10−4 | *** | −1.57 | ↔ | 8.47 × 10−3 | ** | |
| OPG | 1.03 | ↔ | 9.33 × 10−1 | 3.66 | ↑ | 2.55 × 10−3 | ** | ||
| OPN | −2.72 | ↓ | 3.58 × 10−4 | *** | −3.8 | ↓ | 6.58 × 10−4 | *** | |
| RANKL | −1.22 | ↔ | 2.72 × 10−1 | 1.58 | ↔ | 3.01 × 10−1 | |||
| RUNX2 | −1.04 | ↔ | 9.46 × 10−1 | 1.11 | ↔ | 2.58 × 10−1 | |||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deing, A.; Ebel, T.; Willumeit-Römer, R.; Luthringer, B.J.C. Comparison of the Influence of Phospholipid-Coated Porous Ti-6Al-4V Material on the Osteosarcoma Cell Line Saos-2 and Primary Human Bone Derived Cells. Metals 2016, 6, 66. https://doi.org/10.3390/met6030066
Deing A, Ebel T, Willumeit-Römer R, Luthringer BJC. Comparison of the Influence of Phospholipid-Coated Porous Ti-6Al-4V Material on the Osteosarcoma Cell Line Saos-2 and Primary Human Bone Derived Cells. Metals. 2016; 6(3):66. https://doi.org/10.3390/met6030066
Chicago/Turabian StyleDeing, Axel, Thomas Ebel, Regine Willumeit-Römer, and Bérengère J.C. Luthringer. 2016. "Comparison of the Influence of Phospholipid-Coated Porous Ti-6Al-4V Material on the Osteosarcoma Cell Line Saos-2 and Primary Human Bone Derived Cells" Metals 6, no. 3: 66. https://doi.org/10.3390/met6030066
APA StyleDeing, A., Ebel, T., Willumeit-Römer, R., & Luthringer, B. J. C. (2016). Comparison of the Influence of Phospholipid-Coated Porous Ti-6Al-4V Material on the Osteosarcoma Cell Line Saos-2 and Primary Human Bone Derived Cells. Metals, 6(3), 66. https://doi.org/10.3390/met6030066