A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds
Abstract
:1. Introduction
2. Cross-β Aggregation and Amyloids
3. Fungal Adhesins, Amyloid Fibrils, and Cross-β Aggregates
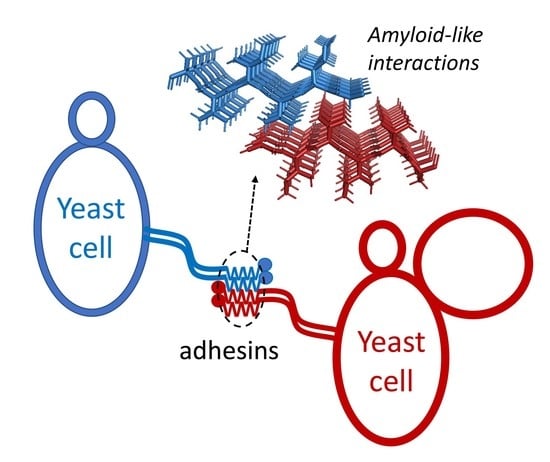
4. Cross-β Bonding in Trans
5. Biogenesis of Cross-β Bonds in Trans
6. Other Adhesins
7. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dehullu, J.; Vorholt, J.A.; Lipke, P.N.; Dufrene, Y.F. Fluidic Force Microscopy Captures Amyloid Bonds between Microbial Cells. Trends Microbiol. 2019, 27, 728–730. [Google Scholar] [CrossRef]
- Dehullu, J.; Valotteau, C.; Herman-Bausier, P.; Garcia-Sherman, M.; Mittelviefhaus, M.; Vorholt, J.A.; Lipke, P.N.; Dufrene, Y.F. Fluidic Force Microscopy Demonstrates That Homophilic Adhesion by Candida albicans Als Proteins Is Mediated by Amyloid Bonds between Cells. Nano Lett. 2019, 19, 3846–3853. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Shewmaker, F.; McGlinchey, R.P.; Wickner, R.B. Structural insights into functional and pathological amyloid. J. Biol. Chem. 2011, 286, 16533–16540. [Google Scholar] [CrossRef] [Green Version]
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like beta-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2017, 82, e0035-17. [Google Scholar] [CrossRef]
- Lipke, P.N. What We Do Not Know about Fungal Cell Adhesion Molecules. J. Fungi 2018, 4, 59. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.X.; Joseph, I.G.; Huang, A.; Jackson, D.N.; Lipke, P.N. Quantitative Analyses of Force-Induced Amyloid Formation in Candida albicans Als5p: Activation by Standard Laboratory Procedures. PLoS ONE 2015, 10, e0129152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formosa, C.; Schiavone, M.; Boisrame, A.; Richard, M.L.; Duval, R.E.; Dague, E. Multiparametric imaging of adhesive nanodomains at the surface of Candida albicans by atomic force microscopy. Nanomedicine 2015, 11, 57–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [Green Version]
- Romling, U.; Kjelleberg, S.; Normark, S.; Nyman, L.; Uhlin, B.E.; Akerlund, B. Microbial biofilm formation: A need to act. J. Intern. Med. 2014, 276, 98–110. [Google Scholar] [CrossRef]
- Blankenship, J.R.; Mitchell, A.P. How to build a biofilm: A fungal perspective. Curr. Opin. Microbiol. 2006, 9, 588–594. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Park, J.; Kahng, B.; Hwang, W. Thermodynamic selection of steric zipper patterns in the amyloid cross-beta spine. PLoS Comput. Biol. 2009, 5, e1000492. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, C.; Pappu, R.V.; Taylor, J.P. Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science 2020, 370, 56–60. [Google Scholar] [CrossRef]
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [Green Version]
- Pinney, J.H.; Hawkins, P.N. Amyloidosis. Ann. Clin. Biochem. 2012, 49, 229–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gerven, N.; Van der Verren, S.E.; Reiter, D.M.; Remaut, H. The Role of Functional Amyloids in Bacterial Virulence. J. Mol. Biol. 2018, 430, 3657–3684. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in Nature, diverse in purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar] [CrossRef]
- Blanco, L.P.; Evans, M.L.; Smith, D.R.; Badtke, M.P.; Chapman, M.R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 2012, 20, 66–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.; Lipke, P.; Klotz, S. Pathogenic microbial amyloids: Their function and the host response. OA Microbiol. 2013, 1, 2. [Google Scholar]
- Sergeeva, A.V.; Galkin, A.P. Functional amyloids of eukaryotes: Criteria, classification, and biological significance. Curr. Genet. 2020, 66, 849–866. [Google Scholar] [CrossRef]
- Taylor, J.D.; Matthews, S.J. New insight into the molecular control of bacterial functional amyloids. Front. Cell. Infect. Microbiol. 2015, 5, 33. [Google Scholar] [CrossRef]
- Jain, N.; Chapman, M.R. Bacterial functional amyloids: Order from disorder. Biochim. Biophys. Acta BBA Proteins Proteom. 2019, 1867, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Vad, B.S.; Dueholm, M.S.; Christiansen, G.; Nilsson, M.; Tolker-Nielsen, T.; Nielsen, P.H.; Meyer, R.L.; Otzen, D.E. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 2015, 6, 1099. [Google Scholar] [CrossRef] [Green Version]
- Dueholm, M.S.; Petersen, S.V.; Sonderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef] [Green Version]
- Watt, B.; van Niel, G.; Raposo, G.; Marks, M.S. PMEL: A pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res. 2013, 26, 300–315. [Google Scholar] [CrossRef] [Green Version]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar] [CrossRef]
- Santos, J.; Pujols, J.; Pallares, I.; Iglesias, V.; Ventura, S. Computational prediction of protein aggregation: Advances in proteomics, conformation-specific algorithms and biotechnological applications. Comput. Struct. Biotechnol. J. 2020, 18, 1403–1413. [Google Scholar] [CrossRef]
- Frousios, K.K.; Iconomidou, V.A.; Karletidi, C.M.; Hamodrakas, S.J. Amyloidogenic determinants are usually not buried. BMC Struct. Biol. 2009, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Ramsook, C.B.; Tan, C.; Garcia, M.C.; Fung, R.; Soybelman, G.; Henry, R.; Litewka, A.; O’Meally, S.; Otoo, H.N.; Khalaf, R.A.; et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 2010, 9, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Younes, S.; Bahnan, W.; Dimassi, H.I.; Khalaf, R.A. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol. Res. 2011, 166, 430–436. [Google Scholar] [CrossRef]
- Garcia, M.C.; Lee, J.T.; Ramsook, C.B.; Alsteens, D.; Dufrene, Y.F.; Lipke, P.N. A role for amyloid in cell aggregation and biofilm formation. PLoS ONE 2011, 6, e17632. [Google Scholar] [CrossRef] [PubMed]
- Gaur, N.K.; Klotz, S.A. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology 2004, 150, 277–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, V.; Herman-Bausier, P.; Shaw, C.; Conrad, K.A.; Garcia-Sherman, M.C.; Draghi, J.; Dufrene, Y.F.; Lipke, P.N.; Rauceo, J.M. An Amyloid Core Sequence in the Major Candida albicans Adhesin Als1p Mediates Cell-Cell Adhesion. MBio 2019, 10, e01766-19. [Google Scholar] [CrossRef] [Green Version]
- Buell, A.K. The growth of amyloid fibrils: Rates and mechanisms. Biochem. J. 2019, 476, 2677–2703. [Google Scholar] [CrossRef] [Green Version]
- Gaur, N.K.; Klotz, S.A.; Henderson, R.L. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect. Immun. 1999, 67, 6040–6047. [Google Scholar] [CrossRef] [Green Version]
- Gaur, N.K.; Klotz, S.A. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 1997, 65, 5289–5294. [Google Scholar] [CrossRef] [Green Version]
- Dranginis, A.M.; Rauceo, J.R.; Coronado, J.E.; Lipke, P.N. A biochemical guide to yeast adhesins: Glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyer, L.L. The ALS gene family of Candida albicans. Trends Microbiol. 2001, 9, 176–180. [Google Scholar] [CrossRef]
- Loza, L.; Fu, Y.; Ibrahim, A.S.; Sheppard, D.C.; Filler, S.G.; Edwards, J.E.J., Jr. Functional analysis of the Candida albicans ALS1 gene product. Yeast 2004, 21, 473–482. [Google Scholar] [CrossRef]
- Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Phan, Q.T.; Fu, Y.; Ibrahim, A.S.; Filler, S.G.; Zhang, M.; Waring, A.J.; Edwards, J.E., Jr. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 2004, 279, 30480–30489. [Google Scholar] [CrossRef] [Green Version]
- Goossens, K.V.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; van Eijsden, R.G.; Siewers, V.; et al. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. MBio 2015, 6, e00427-15. [Google Scholar] [CrossRef] [Green Version]
- Willaert, R.G. Adhesins of Yeasts: Protein Structure and Interactions. J. Fungi 2018, 4, 119. [Google Scholar] [CrossRef] [Green Version]
- Verstrepen, K.J.; Derdelinckx, G.; Verachtert, H.; Delvaux, F.R. Yeast flocculation: What brewers should know. Appl. Microbiol. Biotechnol. 2003, 61, 197–205. [Google Scholar] [CrossRef]
- El-Kirat-Chatel, S.; Beaussart, A.; Vincent, S.P.; Abellan Flos, M.; Hols, P.; Lipke, P.N.; Dufrene, Y.F. Forces in yeast flocculation. Nanoscale 2015, 7, 1760–1767. [Google Scholar] [CrossRef] [Green Version]
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstrepen, K.J.; Reynolds, T.B.; Fink, G.R. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2004, 2, 533–540. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.X.; El-Kirat-Chatel, S.; Joseph, I.G.; Jackson, D.N.; Ramsook, C.B.; Dufrene, Y.F.; Lipke, P.N. Force Sensitivity in Saccharomyces cerevisiae Flocculins. Msphere 2016, 1, e00128-16. [Google Scholar] [CrossRef] [Green Version]
- Smukalla, S.; Caldara, M.; Pochet, N.; Beauvais, A.; Guadagnini, S.; Yan, C.; Vinces, M.D.; Jansen, A.; Prevost, M.C.; Latge, J.P.; et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 2008, 135, 726–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stratford, M. Yeast flocculation: Receptor definition by mnn mutants and concanavalin A. Yeast 1992, 8, 635–645. [Google Scholar] [CrossRef]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [Green Version]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2004, 237, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Kraushaar, T.; Bruckner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mosch, H.U.; Essen, L.O. Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-like Adhesin Domain Girdled by Aromatic Bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, K.V.; Willaert, R.G. The N-terminal domain of the Flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2012, 12, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Lo, W.S.; Dranginis, A.M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by S. cerevisiae. Mol. Biol. Cell 1998, 9, 161–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiavone, M.; Sieczkowski, N.; Castex, M.; Dague, E.; Marie Francois, J. Effects of the strain background and autolysis process on the composition and biophysical properties of the cell wall from two different industrial yeasts. FEMS Yeast Res. 2015, 15, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid Structures as Biofilm Matrix Scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [Green Version]
- Di Martino, P. Bap: A New Type of Functional Amyloid. Trends Microbiol. 2016, 24, 682–684. [Google Scholar] [CrossRef]
- Marinelli, P.; Pallares, I.; Navarro, S.; Ventura, S. Dissecting the contribution of Staphylococcus aureus alpha-phenol-soluble modulins to biofilm amyloid structure. Sci. Rep. 2016, 6, 34552. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formosa, C.; Lachaize, V.; Gales, C.; Rols, M.P.; Martin-Yken, H.; Francois, J.M.; Duval, R.E.; Dague, E. Mapping HA-tagged protein at the surface of living cells by atomic force microscopy. J. Mol. Recognit. 2015, 28, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ryzhova, T.A.; Sopova, J.V.; Zadorsky, S.P.; Siniukova, V.A.; Sergeeva, A.V.; Galkina, S.A.; Nizhnikov, A.A.; Shenfeld, A.A.; Volkov, K.V.; Galkin, A.P. Screening for amyloid proteins in the yeast proteome. Curr. Genet. 2018, 64, 469–478. [Google Scholar] [CrossRef]
- Nizhnikov, A.A.; Alexandrov, A.I.; Ryzhova, T.A.; Mitkevich, O.V.; Dergalev, A.A.; Ter-Avanesyan, M.D.; Galkin, A.P. Proteomic screening for amyloid proteins. PLoS ONE 2014, 9, e116003. [Google Scholar] [CrossRef] [Green Version]
- Kalebina, T.S.; Plotnikova, T.A.; Gorkovskii, A.A.; Selyakh, I.O.; Galzitskaya, O.V.; Bezsonov, E.E.; Gellissen, G.; Kulaev, I.S. Amyloid-like properties of Saccharomyces cerevisiae cell wall glucantransferase Bgl2p: Prediction and experimental evidences. Prion 2008, 2, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Selivanova, O.M.; Glyakina, A.V.; Gorbunova, E.Y.; Mustaeva, L.G.; Suvorina, M.Y.; Grigorashvili, E.I.; Nikulin, A.D.; Dovidchenko, N.V.; Rekstina, V.V.; Kalebina, T.S.; et al. Structural model of amyloid fibrils for amyloidogenic peptide from Bgl2p-glucantransferase of S. cerevisiae cell wall and its modifying analog. New morphology of amyloid fibrils. Biochim. Biophys. Acta BBA Proteins Proteom. 2016, 1864, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Garcia, M.C.; Alsteens, D.; Ramsook, C.B.; Klotz, S.A.; Dufrene, Y.F. Strengthening relationships: Amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012, 20, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.C.; Schauenburg, L.; Thompson-Steckel, G.; Dunsing, V.; Kaden, D.; Voigt, P.; Schaefer, M.; Chiantia, S.; Kennedy, T.E.; Multhaup, G. Amyloid precursor-like protein 1 (APLP1) exhibits stronger zinc-dependent neuronal adhesion than amyloid precursor protein and APLP2. J. Neurochem. 2016, 137, 266–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipke, P.N.; Mathelié-Guinlet, M.; Viljoen, A.; Dufrêne, Y.F. A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds. Pathogens 2021, 10, 1013. https://doi.org/10.3390/pathogens10081013
Lipke PN, Mathelié-Guinlet M, Viljoen A, Dufrêne YF. A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds. Pathogens. 2021; 10(8):1013. https://doi.org/10.3390/pathogens10081013
Chicago/Turabian StyleLipke, Peter N., Marion Mathelié-Guinlet, Albertus Viljoen, and Yves F. Dufrêne. 2021. "A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds" Pathogens 10, no. 8: 1013. https://doi.org/10.3390/pathogens10081013
APA StyleLipke, P. N., Mathelié-Guinlet, M., Viljoen, A., & Dufrêne, Y. F. (2021). A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds. Pathogens, 10(8), 1013. https://doi.org/10.3390/pathogens10081013