Antiviral Agents against Flavivirus Protease: Prospect and Future Direction
Abstract
:1. Introduction
2. Flavivirus Genome Organization
3. Flavivirus Protease
4. Structural Insight of NS2B-NS3 Protease
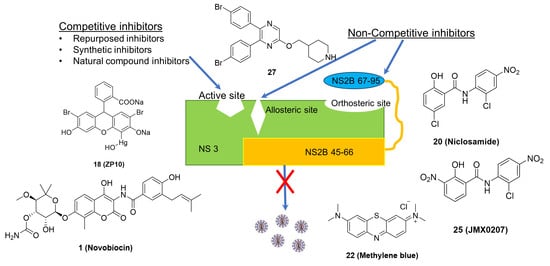
5. NS2B-NS3 Protease Inhibitors
5.1. Competitive Inhibitors
5.1.1. Repurposed Inhibitors
5.1.2. Synthetic Inhibitors
5.1.3. Natural Compound Inhibitors
5.2. Non-Competitive Inhibitors
6. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- Best, S.M. Flaviviruses. Curr. Biol. 2016, 26, R1258–R1260. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, J.; Li, H. Flavivirus NS2B/NS3 Protease: Structure, Function, and Inhibition. In Viral Proteases and Their Inhibitors; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 163–188. [Google Scholar]
- Brecher, M.; Zhang, J.; Li, H. The flavivirus protease as a target for drug discovery. Virol. Sin. 2013, 28, 326–336. [Google Scholar] [CrossRef] [Green Version]
- Kayesh, M.E.H.; Tsukiyama-Kohara, K. Mammalian animal models for dengue virus infection: A recent overview. Arch. Virol. 2021, 167, 31–44. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue/dengue haemorrhagic fever: History and current status. Novartis Found. Symp. 2006, 277, 3–16; discussion 16–22, 71–13, 251–253. [Google Scholar] [CrossRef]
- Pfeffer, M.; Dobler, G. Emergence of zoonotic arboviruses by animal trade and migration. Parasites Vectors 2010, 3, 35. [Google Scholar] [CrossRef] [Green Version]
- Enserink, M. INFECTIOUS DISEASES. An obscure mosquito-borne disease goes global. Science 2015, 350, 1012–1013. [Google Scholar] [CrossRef]
- Holbrook, M. Historical Perspectives on Flavivirus Research. Viruses 2017, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.S.; Gubler, D.J.; Petersen, L.R. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004, 10, S98–S109. [Google Scholar] [CrossRef]
- Roehrig, J. West Nile Virus in the United States—A Historical Perspective. Viruses 2013, 5, 3088–3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Brecher, M.; Li, Z.; Liu, B.; Zhang, J.; Koetzner, C.A.; Alifarag, A.; Jones, S.A.; Lin, Q.; Kramer, L.D.; Li, H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017, 13, e1006411. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Mallewa, M. Dengue and other emerging flaviviruses. J. Infect. 2001, 42, 104–115. [Google Scholar] [CrossRef]
- Norshidah, H.; Vignesh, R.; Lai, N.S. Updates on Dengue Vaccine and Antiviral: Where Are We Heading? Molecules 2021, 26, 6768. [Google Scholar] [CrossRef]
- Meganck, R.M.; Baric, R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021, 27, 401–410. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, M.; Cao, J.; Shen, J.; Zhou, X.; Wang, D.; Cao, J. Flavivirus: From Structure to Therapeutics Development. Life 2021, 11, 615. [Google Scholar] [CrossRef]
- Gebhard, L.G.; Filomatori, C.V.; Gamarnik, A.V. Functional RNA Elements in the Dengue Virus Genome. Viruses 2011, 3, 1739–1756. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Jia, R.; Wang, M.; Yin, Z.; Cheng, A. Structure and function of capsid protein in flavivirus infection and its applications in the development of vaccines and therapeutics. Vet. Res. 2021, 52, 98. [Google Scholar] [CrossRef]
- Obi, J.O.; Gutiérrez-Barbosa, H.; Chua, J.V.; Deredge, D.J. Current Trends and Limitations in Dengue Antiviral Research. Trop. Med. Infect. Dis 2021, 6, 180. [Google Scholar] [CrossRef]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.A.; et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir. Res. 2010, 87, 125–148. [Google Scholar] [CrossRef]
- Natarajan, S. NS3 protease from flavivirus as a target for designing antiviral inhibitors against dengue virus. Genet. Mol. Biol. 2010, 33, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Assenberg, R.; Mastrangelo, E.; Walter, T.S.; Verma, A.; Milani, M.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Mancini, E.J. Crystal structure of a novel conformational state of the flavivirus NS3 protein: Implications for polyprotein processing and viral replication. J. Virol. 2009, 83, 12895–12906. [Google Scholar] [CrossRef] [Green Version]
- Shi, P.Y.; Kauffman, E.B.; Ren, P.; Felton, A.; Tai, J.H.; Dupuis, A.P., 2nd; Jones, S.A.; Ngo, K.A.; Nicholas, D.C.; Maffei, J.; et al. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 2001, 39, 1264–1271. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Yang, K.; Wu, C.; Chen, C.; Hu, C.; Buzovetsky, O.; Wang, Z.; Ji, X.; Xiong, Y.; Yang, H. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 2016, 26, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Chambers, T.J.; Weir, R.C.; Grakoui, A.; McCourt, D.W.; Bazan, J.F.; Fletterick, R.J.; Rice, C.M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc. Natl. Acad. Sci. USA 1990, 87, 8898–8902. [Google Scholar] [CrossRef] [Green Version]
- Löhr, K.; Knox, J.E.; Phong, W.Y.; Ma, N.L.; Yin, Z.; Sampath, A.; Patel, S.J.; Wang, W.L.; Chan, W.L.; Rao, K.R.R.; et al. Yellow fever virus NS3 protease: Peptide-inhibition studies. J. Gen. Virol. 2007, 88, 2223–2227. [Google Scholar] [CrossRef]
- Chappell, K.J.; Nall, T.A.; Stoermer, M.J.; Fang, N.X.; Tyndall, J.D.; Fairlie, D.P.; Young, P.R. Site-directed mutagenesis and kinetic studies of the West Nile Virus NS3 protease identify key enzyme-substrate interactions. J. Biol. Chem. 2005, 280, 2896–2903. [Google Scholar] [CrossRef] [Green Version]
- Falgout, B.; Pethel, M.; Zhang, Y.M.; Lai, C.J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 1991, 65, 2467–2475. [Google Scholar] [CrossRef] [Green Version]
- Cahour, A.; Falgout, B.; Lai, C.J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 1992, 66, 1535–1542. [Google Scholar] [CrossRef] [Green Version]
- Jan, L.R.; Yang, C.S.; Trent, D.W.; Falgout, B.; Lai, C.J. Processing of Japanese encephalitis virus non-structural proteins: NS2B-NS3 complex and heterologous proteases. J. Gen. Virol. 1995, 76 Pt 3, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, D.; Selisko, B.; Locatelli, G.A.; Maga, G.; Romette, J.L.; Canard, B. The RNA helicase, nucleotide 5’-triphosphatase, and RNA 5’-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 2004, 328, 208–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Den Elsen, K.; Quek, J.P.; Luo, D. Molecular Insights into the Flavivirus Replication Complex. Viruses 2021, 13, 956. [Google Scholar] [CrossRef] [PubMed]
- Clum, S.; Ebner, K.E.; Padmanabhan, R. Cotranslational Membrane Insertion of the Serine Proteinase Precursor NS2B-NS3(Pro) of Dengue Virus Type 2 Is Required for Efficient in Vitro Processing and Is Mediated through the Hydrophobic Regions of NS2B. J. Biol. Chem. 1997, 272, 30715–30723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.G.; Lescar, J. Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 Å. J. Virol. 2005, 79, 10278–10288. [Google Scholar] [CrossRef] [Green Version]
- Lescar, J.; Luo, D.; Xu, T.; Sampath, A.; Lim, S.P.; Canard, B.; Vasudevan, S.G. Towards the design of antiviral inhibitors against flaviviruses: The case for the multifunctional NS3 protein from Dengue virus as a target. Antivir. Res. 2008, 80, 94–101. [Google Scholar] [CrossRef]
- Yamashita, T.; Unno, H.; Mori, Y.; Tani, H.; Moriishi, K.; Takamizawa, A.; Agoh, M.; Tsukihara, T.; Matsuura, Y. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase/nucleoside triphosphatase at a resolution of 1.8 A. Virology 2008, 373, 426–436. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Gayen, S.; Kang, C.; Joy, J.; Huang, Q.; Chen, A.S.; Wee, J.L.; Ang, M.J.; Lim, H.A.; Hung, A.W.; et al. NMR analysis of a novel enzymatically active unlinked dengue NS2B-NS3 protease complex. J. Biol. Chem. 2013, 288, 12891–12900. [Google Scholar] [CrossRef] [Green Version]
- Shannon, A.E.; Chappell, K.J.; Stoermer, M.J.; Chow, S.Y.; Kok, W.M.; Fairlie, D.P.; Young, P.R. Simultaneous uncoupled expression and purification of the Dengue virus NS3 protease and NS2B co-factor domain. Protein Expr. Purif. 2016, 119, 124–129. [Google Scholar] [CrossRef] [Green Version]
- Phong, W.Y.; Moreland, N.J.; Lim, S.P.; Wen, D.; Paradkar, P.N.; Vasudevan, S.G. Dengue protease activity: The structural integrity and interaction of NS2B with NS3 protease and its potential as a drug target. Biosci. Rep. 2011, 31, 399–409. [Google Scholar] [CrossRef] [Green Version]
- Hilgenfeld, R.; Lei, J.; Zhang, L. The Structure of the Zika Virus Protease, NS2B/NS3(pro). Adv. Exp. Med. Biol. 2018, 1062, 131–145. [Google Scholar] [CrossRef]
- Wu, C.F.; Wang, S.H.; Sun, C.M.; Hu, S.T.; Syu, W.J. Activation of dengue protease autocleavage at the NS2B-NS3 junction by recombinant NS3 and GST-NS2B fusion proteins. J. Virol. Methods 2003, 114, 45–54. [Google Scholar] [CrossRef]
- Felicetti, T.; Manfroni, G.; Cecchetti, V.; Cannalire, R. Broad-Spectrum Flavivirus Inhibitors: A Medicinal Chemistry Point of View. ChemMedChem 2020, 15, 2391–2419. [Google Scholar] [CrossRef]
- Wensing, A.M.; van Maarseveen, N.M.; Nijhuis, M. Fifteen years of HIV Protease Inhibitors: Raising the barrier to resistance. Antivir. Res. 2010, 85, 59–74. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Rahman, H.M.; Al-karamany, G.S.; El-Koussi, N.A.; Youssef, A.F.; Kiso, Y. HIV protease inhibitors: Peptidomimetic drugs and future perspectives. Curr. Med. Chem. 2002, 9, 1905–1922. [Google Scholar] [CrossRef]
- Wyles, D.L. Antiviral Resistance and the Future Landscape of Hepatitis C Virus Infection Therapy. J. Infect. Dis. 2013, 207, S33–S39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X. Direct anti-HCV agents. Acta Pharm. Sin. B 2016, 6, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Meewan, I.; Zhang, X.; Roy, S.; Ballatore, C.; O’Donoghue, A.J.; Schooley, R.T.; Abagyan, R. Discovery of New Inhibitors of Hepatitis C Virus NS3/4A Protease and Its D168A Mutant. ACS Omega 2019, 4, 16999–17008. [Google Scholar] [CrossRef]
- Ali, A.; Aydin, C.; Gildemeister, R.; Romano, K.P.; Cao, H.; Ozen, A.; Soumana, D.; Newton, A.; Petropoulos, C.J.; Huang, W.; et al. Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chem. Biol. 2013, 8, 1469–1478. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.H.; Nalivaika, E.A.; Prachanronarong, K.L.; Yilmaz, N.K.; Schiffer, C.A. Dengue Protease Substrate Recognition: Binding of the Prime Side. ACS Infect. Dis. 2016, 2, 734–743. [Google Scholar] [CrossRef] [Green Version]
- Choksupmanee, O.; Hodge, K.; Katzenmeier, G.; Chimnaronk, S. Structural Platform for the Autolytic Activity of an Intact NS2B–NS3 Protease Complex from Dengue Virus. Biochemistry 2012, 51, 2840–2851. [Google Scholar] [CrossRef]
- Huang, Q.; Li, Q.; Joy, J.; Chen, A.S.; Ruiz-Carrillo, D.; Hill, J.; Lescar, J.; Kang, C. Lyso-myristoyl phosphatidylcholine micelles sustain the activity of Dengue non-structural (NS) protein 3 protease domain fused with the full-length NS2B. Protein Expr. Purif. 2013, 92, 156–162. [Google Scholar] [CrossRef]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef]
- Noble, C.G.; Seh, C.C.; Chao, A.T.; Shi, P.Y. Ligand-Bound Structures of the Dengue Virus Protease Reveal the Active Conformation. J. Virol. 2012, 86, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Hale, B.G.; Randall, R.E.; Ortín, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef]
- Niyomrattanakit, P.; Winoyanuwattikun, P.; Chanprapaph, S.; Angsuthanasombat, C.; Panyim, S.; Katzenmeier, G. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J. Virol. 2004, 78, 13708–13716. [Google Scholar] [CrossRef] [Green Version]
- Boldescu, V.; Behnam, M.A.M.; Vasilakis, N.; Klein, C.D. Broad-spectrum agents for flaviviral infections: Dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017, 16, 565–586. [Google Scholar] [CrossRef] [Green Version]
- Nitsche, C. Strategies Towards Protease Inhibitors for Emerging Flaviviruses. Adv. Exp. Med. Biol. 2018, 1062, 175–186. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, J.F.; den-Haan, H.; Chik, K.K.; Zhang, A.J.; Chan, C.C.; Poon, V.K.; Yip, C.C.; Mak, W.W.; Zhu, Z.; et al. Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antivir. Res. 2017, 145, 33–43. [Google Scholar] [CrossRef]
- Pathak, N.; Lai, M.L.; Chen, W.Y.; Hsieh, B.W.; Yu, G.Y.; Yang, J.M. Pharmacophore anchor models of flaviviral NS3 proteases lead to drug repurposing for DENV infection. BMC Bioinform. 2017, 18, 548. [Google Scholar] [CrossRef] [Green Version]
- Rassias, G.; Zogali, V.; Swarbrick, C.M.D.; Ki Chan, K.W.; Chan, S.A.; Gwee, C.P.; Wang, S.; Kaplanai, E.; Canko, A.; Kiousis, D.; et al. Cell-active carbazole derivatives as inhibitors of the zika virus protease. Eur. J. Med. Chem. 2019, 180, 536–545. [Google Scholar] [CrossRef]
- Abrams, R.P.M.; Yasgar, A.; Teramoto, T.; Lee, M.H.; Dorjsuren, D.; Eastman, R.T.; Malik, N.; Zakharov, A.V.; Li, W.; Bachani, M.; et al. Therapeutic candidates for the Zika virus identified by a high-throughput screen for Zika protease inhibitors. Proc. Natl. Acad. Sci. USA 2020, 117, 31365–31375. [Google Scholar] [CrossRef]
- Akaberi, D.; Båhlström, A.; Chinthakindi, P.K.; Nyman, T.; Sandström, A.; Järhult, J.D.; Palanisamy, N.; Lundkvist, Å.; Lennerstrand, J. Targeting the NS2B-NS3 protease of tick-borne encephalitis virus with pan-flaviviral protease inhibitors. Antivir. Res. 2021, 190, 105074. [Google Scholar] [CrossRef]
- Wu, D.W.; Mao, F.; Ye, Y.; Li, J.; Xu, C.L.; Luo, X.M.; Chen, J.; Shen, X. Policresulen, a novel NS2B/NS3 protease inhibitor, effectively inhibits the replication of DENV2 virus in BHK-21 cells. Acta Pharm. Sin. 2015, 36, 1126–1136. [Google Scholar] [CrossRef] [Green Version]
- Takagi, Y.; Matsui, K.; Nobori, H.; Maeda, H.; Sato, A.; Kurosu, T.; Orba, Y.; Sawa, H.; Hattori, K.; Higashino, K.; et al. Discovery of novel cyclic peptide inhibitors of dengue virus NS2B-NS3 protease with antiviral activity. Bioorg. Med. Chem. Lett. 2017, 27, 3586–3590. [Google Scholar] [CrossRef]
- Behnam, M.A.; Graf, D.; Bartenschlager, R.; Zlotos, D.P.; Klein, C.D. Discovery of Nanomolar Dengue and West Nile Virus Protease Inhibitors Containing a 4-Benzyloxyphenylglycine Residue. J. Med. Chem. 2015, 58, 9354–9370. [Google Scholar] [CrossRef]
- Lin, K.H.; Ali, A.; Rusere, L.; Soumana, D.I.; Kurt Yilmaz, N.; Schiffer, C.A. Dengue Virus NS2B/NS3 Protease Inhibitors Exploiting the Prime Side. J. Virol. 2017, 91, e00045-17. [Google Scholar] [CrossRef] [Green Version]
- Cabarcas-Montalvo, M.; Maldonado-Rojas, W.; Montes-Grajales, D.; Bertel-Sevilla, A.; Wagner-Döbler, I.; Sztajer, H.; Reck, M.; Flechas-Alarcon, M.; Ocazionez, R.; Olivero-Verbel, J. Discovery of antiviral molecules for dengue: In silico search and biological evaluation. Eur. J. Med. Chem. 2016, 110, 87–97. [Google Scholar] [CrossRef]
- Bastos Lima, A.; Behnam, M.A.; El Sherif, Y.; Nitsche, C.; Vechi, S.M.; Klein, C.D. Dual inhibitors of the dengue and West Nile virus NS2B-NS3 proteases: Synthesis, biological evaluation and docking studies of novel peptide-hybrids. Bioorg. Med. Chem. 2015, 23, 5748–5755. [Google Scholar] [CrossRef]
- Skoreński, M.; Milewska, A.; Pyrć, K.; Sieńczyk, M.; Oleksyszyn, J. Phosphonate inhibitors of West Nile virus NS2B/NS3 protease. J. Enzym. Inhib. Med. Chem. 2019, 34, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Zhou, R.; Huang, C.; Zhang, R.; Wang, J.; Zhang, Y.; Ding, J.; Li, X.; Zhou, J.; Cen, S. Identification of Theaflavin-3,3’-Digallate as a Novel Zika Virus Protease Inhibitor. Front. Pharmacol. 2020, 11, 514313. [Google Scholar] [CrossRef]
- Mueller, N.H.; Yon, C.; Ganesh, V.K.; Padmanabhan, R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int. J. Biochem. Cell Biol. 2007, 39, 606–614. [Google Scholar] [CrossRef]
- Li, Z.; Brecher, M.; Deng, Y.Q.; Zhang, J.; Sakamuru, S.; Liu, B.; Huang, R.; Koetzner, C.A.; Allen, C.A.; Jones, S.A.; et al. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 2017, 27, 1046–1064. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Shi, P.Y.; Li, H.; Zhou, J. Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infect. Dis. 2020, 6, 909–915. [Google Scholar] [CrossRef]
- Shi, Z.; Wei, J.; Deng, X.; Li, S.; Qiu, Y.; Shao, D.; Li, B.; Zhang, K.; Xue, F.; Wang, X.; et al. Nitazoxanide inhibits the replication of Japanese encephalitis virus in cultured cells and in a mouse model. Virol. J. 2014, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, J.F. Nitazoxanide: A first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014, 110, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Lang, Y.; Sakamuru, S.; Samrat, S.; Trudeau, N.; Kuo, L.; Rugenstein, N.; Tharappel, A.; D’Brant, L.; Koetzner, C.A.; et al. Methylene blue is a potent and broad-spectrum inhibitor against Zika virus in vitro and in vivo. Emerg. Microbes Infect. 2020, 9, 2404–2416. [Google Scholar] [CrossRef]
- Li, Z.; Sakamuru, S.; Huang, R.; Brecher, M.; Koetzner, C.A.; Zhang, J.; Chen, H.; Qin, C.F.; Zhang, Q.Y.; Zhou, J.; et al. Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antivir. Res. 2018, 150, 217–225. [Google Scholar] [CrossRef]
- Li, Z.; Xu, J.; Lang, Y.; Wu, X.; Hu, S.; Samrat, S.; Tharappel, A.; Kuo, L.; Butler, D.; Song, Y.; et al. In vitro and in vivo characterization of erythrosin B and derivatives against Zika virus. Acta Pharm. Sin. B 2021. [Google Scholar] [CrossRef]
- Li, Z.; Xu, J.; Lang, Y.; Fan, X.; Kuo, L.; D’Brant, L.; Hu, S.; Samrat, S.K.; Trudeau, N.; Tharappel, A.M.; et al. JMX0207, a Niclosamide Derivative with Improved Pharmacokinetics, Suppresses Zika Virus Infection Both In Vitro and In Vivo. ACS Infect. Dis. 2020, 6, 2616–2628. [Google Scholar] [CrossRef]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef]
- Nie, S.; Yao, Y.; Wu, F.; Wu, X.; Zhao, J.; Hua, Y.; Wu, J.; Huo, T.; Lin, Y.L.; Kneubehl, A.R.; et al. Synthesis, Structure-Activity Relationships, and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Med. Chem. 2021, 64, 2777–2800. [Google Scholar] [CrossRef]
- Batool, F.; Saeed, M.; Saleem, H.N.; Kirschner, L.; Bodem, J. Facile Synthesis and In Vitro Activity of N-Substituted 1,2-Benzisothiazol-3(2H)-ones against Dengue Virus NS2BNS3 Protease. Pathogens 2021, 10, 464. [Google Scholar] [CrossRef]
- Millies, B.; von Hammerstein, F.; Gellert, A.; Hammerschmidt, S.; Barthels, F.; Göppel, U.; Immerheiser, M.; Elgner, F.; Jung, N.; Basic, M.; et al. Proline-Based Allosteric Inhibitors of Zika and Dengue Virus NS2B/NS3 Proteases. J. Med. Chem. 2019, 62, 11359–11382. [Google Scholar] [CrossRef]




| Number (Name) of Compound | Targeted Virus | IC50 (or Ki) (μM) | EC50 (μM) | In Vivo | Reference |
|---|---|---|---|---|---|
| 1 (Novobiocin) | ZIKV | 14.2 ± 1.1 | 42.63 | Yes | [61] |
| 2 (Asunaprevir) | ZIKV | 6.0 | 4.7 | [62] | |
| 3 (Simeprevir) | ZIKV | 2.6 | 0.4 | [62] | |
| 4 (Carbazole-based amidines) | ZIKV | 0.52 | 1.25 | [63] | |
| 5 (MK-591) | ZIKV | 3.0 | 3.1 | [64] | |
| 6 (JNJ-40418677) | ZIKV | 3.9 | 3.2 | [64] | |
| 7 (4-CF3-benzyl ether) | TBEV | 0.92 | [65] | ||
| ZIKV | 1.64 | ||||
| 8 | ZIKV | 0.25 | [65] | ||
| TBEV | 0.97 | ||||
| DENV | 0.05 | 3.4 | |||
| WNV | 0.018 | 15.5 | |||
| 9 | ZIKV | 0.94 | [65] | ||
| TBEV | 3.72 | ||||
| 10 (Policresulen) | DENV-2 | 0.81 | 8.47 | [66] | |
| 11 | DENV-2 | 0.95 | [67,68] | ||
| 12 | DENV | 1.1 | 2.0 | [67] | |
| 13 (PCRARIYGGCA) | DENV-3 | Ki = 2.9 | [69] | ||
| 14 (C30H25NO5) | DENV-2 | 17.46 | 14.9 | [70] | |
| 15 (C34H23NO7S2) | DENV-2 | 9.09 | 11.8 | [70] | |
| 16 (Peptide-hybrid inhibitors based on 2,4-thiazolidinedione scaffold) | WNV | 0.75 | [71] | ||
| DENV | 1.05 | ||||
| 17 (α-aminoalkylphosphonates) | DENV-2 | Ki = 0.4 | [72] | ||
| 18 (Theaflavin-3,3′-digallate (ZP10) | ZIKV | 7.65 | 3 | [73] |
| Number (Name) of Compound | Targeted Virus | IC50 (μM) | EC50 (μM) | In Vivo | Reference |
|---|---|---|---|---|---|
| 19 (Temoporfin) | DENV-2 | 1.1 ± 0.1 | 0.020 | [75] | |
| ZIKV | 0.024 | Yes | |||
| WNV | 0.010 | ||||
| JEV | 0.011 | ||||
| YFV | 0.006 | ||||
| 20 (Niclosamide) | DENV-2 | 12.3 ± 0.6 | 0.55 | [75,76] | |
| ZIKV | 0.48 | ||||
| WNV | 0.54 | ||||
| JEV | 1.02 | ||||
| YFV | 0.84 | ||||
| 21 (Nitazoxanide) | DENV-2 | 15.9 ± 0.9 | [75,77] | ||
| ZIKV | 1.48 | ||||
| JEV | 0.39 | Yes | |||
| (Tizoxanide) | DENV-2 | 0.38 | [78] | ||
| YFV | 0.23 | ||||
| 22 (Methylene blue) | DENV-2 | 8.9 | 0.36 | [79] [79] | |
| ZIKV | 0.087–0.2 | Yes | |||
| 23 (Erythrosin B) | DENV-2 | 1.9 | 1.2 | [80] | |
| ZIKV | 1.7 | 0.62 | Yes | ||
| WNV | 0.66 | ||||
| JEV | 0.35 | ||||
| YFV | 0.57 | ||||
| 24 (JMX0902) | ZIKV | 2.6 | 0.3 | [81] | |
| 25 (JMX0207) | DENV-2 | 8.2 | 0.31 | [82] | |
| ZIKV | 0.3 | Yes | |||
| 26 (NSC135618) | DENV-2 | 1.8 | 0.81 | [14] | |
| ZIKV | 1.0 | ||||
| WNV | 1.27 | ||||
| YFV | 0.28 | ||||
| 27 | ZIKV | 21.7 | [83] | ||
| 28 | ZIKV | 3.1 | [83] | ||
| 29 | ZIKV | 0.20 ± 0.01 | EC68 0.3 or 0.6 | Yes | [83,84] |
| DENV-2 | 0.59 ± 0.02 | ||||
| DENV-3 | 0.52 ± 0.06 | ||||
| WNV | 0.78 ± 0.02 | ||||
| 30 | ZIKV | 0.13 | EC68 0.6 | [84] | |
| DENV-2 | 2.4 | ||||
| WNV | 0.82 | ||||
| 31 | DENV-2 | 2.56 ± 1.03 | <30 | [85] | |
| 32 | DENV-2 | 2.01± 0.98 | <30 | [85] | |
| 33 | DENV-2 | 5.28 ± 1.89 | <30 | [85] | |
| (R)-34 | DENV-2 | 0.32 | <3 | [86] | |
| (R)-35 | DENV-2 | 0.51 | [86] | ||
| (S)-35 | DENV-2 | 0.58 | [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samrat, S.K.; Xu, J.; Li, Z.; Zhou, J.; Li, H. Antiviral Agents against Flavivirus Protease: Prospect and Future Direction. Pathogens 2022, 11, 293. https://doi.org/10.3390/pathogens11030293
Samrat SK, Xu J, Li Z, Zhou J, Li H. Antiviral Agents against Flavivirus Protease: Prospect and Future Direction. Pathogens. 2022; 11(3):293. https://doi.org/10.3390/pathogens11030293
Chicago/Turabian StyleSamrat, Subodh K., Jimin Xu, Zhong Li, Jia Zhou, and Hongmin Li. 2022. "Antiviral Agents against Flavivirus Protease: Prospect and Future Direction" Pathogens 11, no. 3: 293. https://doi.org/10.3390/pathogens11030293
APA StyleSamrat, S. K., Xu, J., Li, Z., Zhou, J., & Li, H. (2022). Antiviral Agents against Flavivirus Protease: Prospect and Future Direction. Pathogens, 11(3), 293. https://doi.org/10.3390/pathogens11030293