Optimization of a Protocol for Launching Grapevine Infection with the Biologically Active cDNA Clones of a Virus
Abstract
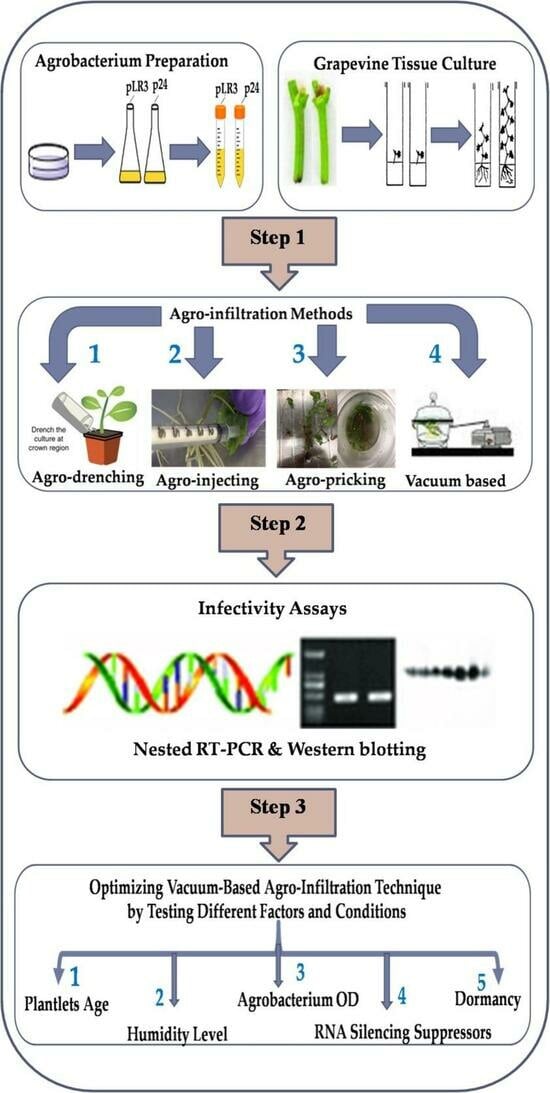
:1. Introduction
2. Material and Methods
2.1. Establishment of Grapevine Tissue Culture
2.2. Agrobacterium Preparation
2.3. Four Methods Applied in Agro-Inoculation
2.3.1. Vacuum-Based Agro-Infiltration
2.3.2. Agro-Pricking
2.3.3. Agro-Drenching
2.3.4. Agro-Injection
2.4. The Effects of Various Factors on the Survival and Infectivity Rate Using Vacuum-Based Agro-Infiltration Technique
2.4.1. Age and the Cultivar of Tissue-Cultured Plantlets
2.4.2. Impacts of Humidity Level
2.4.3. Effects of Vacuum Duration and Agrobacterial Density (OD600) on Plantlet Survival and Infectivity
2.4.4. Effects of Various RSSs on Infectivity
2.4.5. Impact of Dormancy Treatment on the Infection Rates and Viral Titer
2.5. Infectivity Assays
2.5.1. Total RNA Isolation, RT-PCR, Nested RT-PCR, and RT-qPCR
2.5.2. Western Blotting
3. Results
3.1. Vacuum-Based Agro-Infiltration Is the Best Approach for Agro-Infection
3.2. Impacts of Age and the Cultivar of Grapevine Plantlets on Survival and Infectivity Rates
3.3. The Effects of Humidity Control on Plantlet Survival and Infectivity
3.4. Effects of Vacuum Treatment Duration and Density of Agrobacterium on Infectivity Rates
3.5. Effects of Co-Infiltration with Virus RSSs on Infectivity
3.6. Effects of Dormancy Treatment on Infection Rate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martelli, G.P. An overview on grapevine viruses, viroids, and the diseases they cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 31–46. [Google Scholar]
- Martelli, G.P. Where Grapevine Virology Is Heading To. In Proceedings of the 19th Congress of ICVG, Santiago, Chile, 9–12 April 2018; pp. 10–15. [Google Scholar]
- Fuchs, M. Grapevine viruses: A multitude of diverse species with simple but overall, poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Crnogorac, A.; Panno, S.; Mandić, A.; Gašpar, M.; Caruso, A.G.; Noris, E.; Davino, S.; Maltic, S. Survey of five major grapevine viruses infecting Blatina and Žilavka cultivars in Bosnia and Herzegovina. PLoS ONE 2021, 16, e0245959. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 3–31. [Google Scholar]
- Saldarelli, P.; Giampetruzzi, A.; Maree, H.J.; Rwahnih, A. High Throughput Sequencing: Advantages beyond Virus Identification; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 625–642. [Google Scholar]
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Maliogka, V.I.; Martelli, G.P.; Fuchs, M.; Katis, N.I. Control of viruses infecting grapevine. Adv. Virus Res. 2015, 91, 175–227. [Google Scholar] [PubMed]
- Al Rwahnih, M.; Dave, A.; Anderson, M.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R. Association of a DNA virus with grapevines affected by red blotch disease in California. Phytopathology 2013, 103, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine red blotch-associated virus is widespread in the United States. Phytopathology 2012, 104, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Poojari, S.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. Incidence, distribution and genetic diversity of grapevine red blotch virus in British Columbia. Can. J. Plant Pathol. 2017, 39, 201–211. [Google Scholar] [CrossRef]
- Saldarelli, P.; Gualandri, V.; Malossini, U.; Glasa, M. Grapevine Pinot Gris Virus; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 351–364. [Google Scholar]
- Giampetruzzi, A.; Roumi, V.; Roberto, R.; Malossini, U.; Yoshikawa, N.; La Notte, P.; Terlizzi, F.; Credi, R.; Saldarelli, P. A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in Cv Pinot gris. Virus Res. 2012, 163, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Kreuze, K.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine leafroll-associated virus-3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine leafroll: A complex viral disease affecting a high-value fruit crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Turpen, T.H.; Turpen, A.M.; Weinzettl, N.; Kumagai, M.H.; Dawson, W.O. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of Tobacco mosaic virus. J. Virol. Methods 1993, 42, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, H.B. Rapid delivery of foreign genes into plants by direct rub-inoculation wh intact plasmid DNA of a Tomato bushy stunt virus gene vector. J. Virol. 1999, 73, 7823–7829. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Bar-Joseph, M.; Mawassi, M.; Albiach-Marti, M.R.; Ayllon, M.A.; Gowda, S.; Hilf, M.E.; Moreno, P.; Garnsey, S.M.; Dawson, W.O. Amplification of Citrus tristeza virus from a cDNA Clone and Infection of Citrus Trees. Virology 2001, 280, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Muruganantham, M.; Moskovitz, Y.; Haviv, S.; Horesh, T.; Fenigstein, A.; Preez, J.; Stephan, D.; Burger, J.T.; Mawassi, M. Grapevine virus A-mediated gene silencing in Nicotiana benthamiana and Vitis vinifera. J. Virol. Methods 2009, 155, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Kurth, E.G.; Peremyslov, V.V.; Prokhnevsky, A.I.; Kasschau, K.D.; Miller, M.; Carrington, J.C.; Dolja, V.V. Virus-derived gene expression and RNA interference vector for grapevine. Virology 2012, 86, 6002–6009. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Venkataraman, S.; Li, C.; Wanga, W.; Dayan-Glick, C.; Mawassi, M. Construction and biological activities of the first infectious cDNA clones of the genus Foveavirus. Virology 2013, 435, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Lovato, A.; Faoro, F.; Gambino, G.; Maffi, D.; Bracale, M.; Polverari, A.; Santi, L. Construction of a synthetic infectious cDNA clone of Grapevine Algerian latent virus (GALV-Nf) and its biological activity in Nicotiana benthamiana and grapevine plants. Virology 2014, 11, 186. [Google Scholar] [CrossRef]
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative Role of Grapevine Red Blotch Virus in Red Blotch Disease. Phythopathology 2018, 108, 902–909. [Google Scholar] [CrossRef]
- Jarugula, S.; Gowda, S.; Dawson, W.O.; Naidu, R. Development of infectious cDNA clones of Grapevine leafroll-associated virus 3 and analyses of the 5′ non-translated region for replication and virion formation. Virology 2018, 523, 89–99. [Google Scholar] [CrossRef]
- Tarquini, G.; Zaina, G.; Ermacora, P.; Amicis, F.D.; Franco-Orozco, B.; Loi, N.; Martini, M.; Bianchi, G.L.; Pagliari, L.; Firrao, G.; et al. Agroinoculation of Grapevine Pinot gris virus in tobacco and grapevine provides insights on viral pathogenesis. PLoS ONE 2019, 14, e0214010. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Rietman, H.; Vleeshouwers, V.G. Agroinfiltration and PVX Agroinfection in Potato and Nicotiana benthamiana. J. Vis. Exp. 2014, 83, e50971. [Google Scholar] [CrossRef]
- Ryu, C.M.; Anand, A.; Kang, L.; Mysore, K.S. Agrodrench: A novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J. 2004, 40, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jelkmann, W. Construction of full-length infectious cDNA clones of Apple chlorotic leaf spot virus and their agroinoculation to woody plants by a novel method of vacuum infiltration. Plant Dis. 2017, 101, 2110–2115. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Shabanian, M.; Moore, C.; Li, C.; Meng, B. Survey for major viruses in commercial Vitis vinifera wine grapes in Ontario. Virol. J. 2018, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Seaton, K. Semi-sterilized tissue culture for rapid propagation of grapevines (Vitis vinifera L.) using immature cuttings. Hortscience 2014, 49, 949–954. [Google Scholar] [CrossRef]
- Zottini, M.; Barizza, E.; Costa, A.; Formentin, E.; Ruberti, C.; Carimi, F.; Lo Schiavo, F. Agroinfiltration of grapevine leaves for fast transient assays of gene expression and for long-term production of stable transformed cells. Plant Cell Rep. 2008, 27, 845–885. [Google Scholar] [CrossRef]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef]
- Shabanian, M.; Xiao, H.; Meng, B. Seasonal dynamics and tissue distribution of two major viruses associated with grapevine leafroll under cool climate conditions. Eur. J. Plant Pathol. 2020, 158, 1017–1031. [Google Scholar] [CrossRef]
- Liu, Y.P.; Peremyslov, V.V.; Medina, V.; Dolja, V.V. Tandem leader proteases of grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology 2009, 383, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Mikona, C.; Jelkmann, W. Replication of Grapevine leafroll-associated virus-7 (GLRaV-7) by Cuscuta species and its transmission to herbaceous plants. Plant Dis. 2010, 94, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.G.; Fuchs, M. Herbaceous plant hosts as supermodels for grapevine viruses: A historical perspective. J. Plant Pathol. 2022, 27, 1–30. [Google Scholar] [CrossRef]
- Grimsley, N.; Hohn, B.; Hohn, T.; Walden, R. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. USA 1986, 83, 3282–3286. [Google Scholar] [CrossRef] [PubMed]
- Chiba, M.; Reed, J.C.; Prokhnevsky, A.I.; Chapman, E.J.; Mawassi, M.; Koonin, E.V.; Carrington, J.C.; Dolja, V.V. Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 2006, 346, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Thompson, J.; Fuchs, M.; Perry, K.L. Complete genome sequence of a new circular DNA virus from grapevine. J. Virol. 2012, 86, 7715. [Google Scholar] [CrossRef]








| Inoculation Method | Vacuum-Based Agro-Infiltration | Agro-Pricking | Agro-Drenching | Agro-Injection | ||||
|---|---|---|---|---|---|---|---|---|
| Cultivar of Plantlets | SY | CF | SY | CF | SY | CF | SY | CF |
| No. of infiltrated plantlets | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| No. of survivor plantlets | 39 | 38 | 43 | 42 | 46 | 45 | 44 | 43 |
| No. of infected plantlets | 22 | 20 | 7 | 6 | 0 | 0 | 0 | 0 |
| Percentage of infection of survivor plantlets | 56% | 52% | 16% | 14% | 0% | 0% | 0% | 0% |
| Age of Plantlets | 5–7 Weeks Old | 8–11 Weeks Old | 12–16 Weeks Old | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cultivar of Plantlets | SY | CF | CH | SY | CF | CH | SY | CF | CH |
| No. of infiltrated plantlets | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| No. of survivor plantlets | 25 | 21 | 18 | 82 | 78 | 65 | 92 | 88 | 76 |
| No. of infected plantlets | 14 | 11 | 8 | 52 | 46 | 29 | 13 | 10 | 5 |
| Percentage of infection of survivor plantlets | 56% | 52% | 44% | 63% | 58% | 44% | 14% | 11% | 7% |
| Time of Cover Removal | 1 Week | 2 Weeks | 3 Weeks | |||
|---|---|---|---|---|---|---|
| Manner of Cover Removal | Instant | Gradual | Instant | Gradual | Instant | Gradual |
| No. of infiltrated plantlets | 100 | 100 | 100 | 100 | 100 | 100 |
| No. of survivor plantlets | 0 | 6 | 2 | 18 | 36 | 78 |
| No. of infected plantlets | 0 | 0 | 0 | 4 | 11 | 51 |
| Percentage of infection of survivor plantlets | 0% | 0% | 0% | 22% | 30% | 65% |
| Vacuum Duration | 5 min | 10 min | 15 min | |||
|---|---|---|---|---|---|---|
| Cultivar of Plantlets | SY | CF | SY | CF | SY | CF |
| No. of infiltrated plantlets | 50 | 50 | 50 | 50 | 50 | 50 |
| No. of survivorplantlets | 42 | 41 | 39 | 37 | 21 | 18 |
| No. of infected plantlets | 8 | 5 | 26 | 24 | 12 | 11 |
| Percentage of infection of survivor plantlets | 19% | 12% | 66% | 64% | 57% | 61% |
| OD 600 | GLRaV-3 | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|---|
| P24 | 0.5 | 1 | 0.5 | 1 | 0.5 | 1 | |
| Cultivar of Plantlets | SY | SY | SY | SY | SY | SY | |
| No. of infiltrated plantlets | 50 | 50 | 50 | 50 | 50 | 50 | |
| No. of survivor plantlets | 42 | 39 | 38 | 34 | 23 | 18 | |
| No. of infected plantlets | 5 | 9 | 26 | 24 | 15 | 11 | |
| Percentage of infection of survivor plantlets | 12% | 23% | 68% | 70% | 65% | 61% | |
| RNA-Silencing Suppressors | No SRS | HC-Pro | P24 | P19 | TCV | P21 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SY | CF | SY | CF | SY | CF | SY | CF | SY | CF | SY | CF | |
| No. of infiltrated vines | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| No. of survivor vines | 44 | 40 | 42 | 37 | 40 | 39 | 44 | 38 | 39 | 36 | 43 | 38 |
| No. of infected vines | 8 | 9 | 22 | 19 | 20 | 18 | 26 | 23 | 21 | 23 | 17 | 18 |
| Percentage of infection of survivor vines | 18% | 23% | 52% | 51% | 50% | 51% | 59% | 60% | 54% | 64% | 40% | 47% |
| Rank | Based on Infection % of All Plants | Based on Infection % of Survivor Plants | ||
|---|---|---|---|---|
| SY | CF | SY | CF | |
| 1 | P19 (52%) | TCV (46%) | P19 (59%) | TCV (64%) |
| 2 | HC-Pro (44%) | P19 (46%) | TCV (54%) | P19 (60%) |
| 3 | TCV (42%) | HC-Pro (38%) | HC-Pro (52%) | HC-Pro (51%) |
| 4 | P24 (40%) | P24 (36%) | P24 (50%) | P24 (46%) |
| 5 | P21 (34%) | P21 (36%) | P21 (40%) | P21 (47%) |
| No RSS | 16% | 18% | 18% | 23% |
| Duration of Dormancy at 4 °C | No Dormancy Treatment | Single Dormancy | Double Dormancy | |||
|---|---|---|---|---|---|---|
| Cultivar of Plantlets | SY | CF | SY | CF | SY | CF |
| Total no. of infiltrated plantlets | 50 | 50 | 50 | 50 | 50 | 50 |
| No. of plants that survived dormancy treatment | 50 | 50 | 47 | 45 | 29 | 27 |
| No. of infected plantlets prior to dormancy | 25 | 25 | 25 | 25 | 25 | 25 |
| No. of plantlets that tested positive after dormancy | 26 | 27 | 32 | 33 | 11 | 9 |
| Percentage of infection of survivor plantlets | 52% | 54% | 68% | 73% | 37% | 33% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shabanian, M.; Li, C.; Ebadi, A.; Dolja, V.; Meng, B. Optimization of a Protocol for Launching Grapevine Infection with the Biologically Active cDNA Clones of a Virus. Pathogens 2023, 12, 1314. https://doi.org/10.3390/pathogens12111314
Shabanian M, Li C, Ebadi A, Dolja V, Meng B. Optimization of a Protocol for Launching Grapevine Infection with the Biologically Active cDNA Clones of a Virus. Pathogens. 2023; 12(11):1314. https://doi.org/10.3390/pathogens12111314
Chicago/Turabian StyleShabanian, Mehdi, Caihong Li, Ali Ebadi, Valerian Dolja, and Baozhong Meng. 2023. "Optimization of a Protocol for Launching Grapevine Infection with the Biologically Active cDNA Clones of a Virus" Pathogens 12, no. 11: 1314. https://doi.org/10.3390/pathogens12111314
APA StyleShabanian, M., Li, C., Ebadi, A., Dolja, V., & Meng, B. (2023). Optimization of a Protocol for Launching Grapevine Infection with the Biologically Active cDNA Clones of a Virus. Pathogens, 12(11), 1314. https://doi.org/10.3390/pathogens12111314