Phylogenetic Investigations of Dengue 2019–2021 Outbreak in Guadeloupe and Martinique Caribbean Islands
Abstract
:1. Introduction
2. Material and Methods
2.1. Ethic Statement
2.2. Human Sample Collection and Dengue Virus Screening
2.3. Viral Genome Sequencing
2.4. Recombination Study
2.5. Phylogenetic Analysis
3. Results
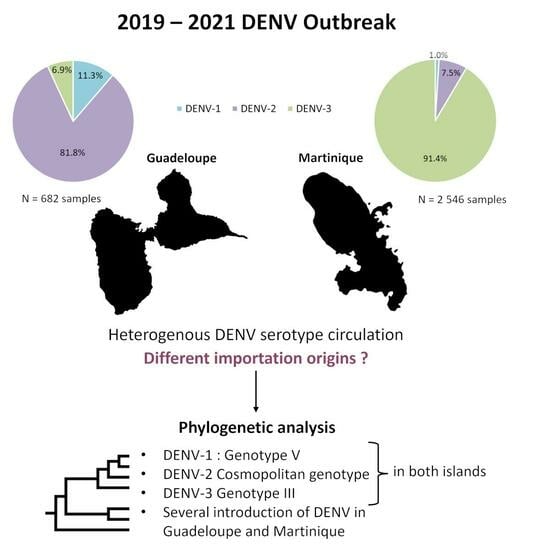
3.1. Heterogeneity of the Main Dengue Virus Serotype Circulation Confirmed for Guadeloupe and Martinique during the 2019–2021 Outbreak
3.2. Two Origins of Introduction of DENV-1 Genotype V in Guadeloupe
3.3. Homogenous Circulation of DENV-2 Cosmopolitan Genotype in Guadeloupe and Martinique Islands
3.4. Two Different DENV-3 Genotype III Introductions in Guadeloupe and Martinique
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Brady, O.J.; Scott, T.W.; Zou, C.; Pigott, D.M.; Duda, K.A.; Bhatt, S.; Katzelnick, L.; Howes, R.E.; Battle, K.E.; et al. Global Spread of Dengue Virus Types: Mapping the 70 Year History. Trends Microbiol. 2014, 22, 138–146. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 14 March 2022).
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The Global Distribution and Burden of Dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Parreira, R.; Sousa, C.A. Dengue Fever in Europe: Could There Be an Epidemic in the Future? Expert. Rev. Anti. Infect. Ther. 2015, 13, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Añez, G.; Rios, M. Dengue in the United States of America: A Worsening Scenario? BioMed Res. Int. 2013, 2013, e678645. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Nakayama, E.; Kotaki, A.; Moi, M.L.; Ikeda, M.; Yagasaki, K.; Saito, Y.; Shibasaki, K.; Saijo, M.; Takasaki, T. Whole Genome Sequencing-Based Molecular Epidemiologic Analysis of Autochthonous Dengue Virus Type 1 Strains Circulating in Japan in 2014. Jpn. J. Infect. Dis. 2017, 70, 45–49. [Google Scholar] [CrossRef]
- Halstead, S.B. Is Dengue Vaccine Protection Possible? Clin. Infect. Dis. 2022, 74, 156–160. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Ooi, E.-E.; Vasudevan, S.G.; Gubler, D.J. Update on Dengue: Epidemiology, Virus Evolution, Antiviral Drugs, and Vaccine Development. Curr. Infect. Dis. Rep. 2010, 12, 157–164. [Google Scholar] [CrossRef]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the Global Spatial Limits of Dengue Virus Transmission by Evidence-Based Consensus. PLoS Neglected Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef]
- Henchal, E.A.; Putnak, J.R. The Dengue Viruses. Clin. Microbiol. Rev. 1990, 3, 376–396. [Google Scholar] [CrossRef]
- Chambers, T.J.; Hahn, C.S.; Galler, R.; Rice, C.M. Flavivirus Genome Organization, Expression, and Replication. Annu. Rev. Microbiol. 1990, 44, 649–688. [Google Scholar] [CrossRef]
- Wang, E.; Ni, H.; Xu, R.; Barrett, A.D.; Watowich, S.J.; Gubler, D.J.; Weaver, S.C. Evolutionary Relationships of Endemic/Epidemic and Sylvatic Dengue Viruses. J. Virol. 2000, 74, 3227–3234. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Vasilakis, N. Molecular Evolution of Dengue Viruses: Contributions of Phylogenetics to Understanding the History and Epidemiology of the Preeminent Arboviral Disease. Infect. Genet. Evol. 2009, 9, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Arriaga, J.B.; Green, A.; James, S.L.; Lau, L.; Montoya, M.; Wang, C.; VanBlargan, L.A.; et al. Dengue Viruses Cluster Antigenically but Not as Discrete Serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef]
- Rico-Hesse, R. Molecular Evolution and Distribution of Dengue Viruses Type 1 and 2 in Nature. Virology 1990, 174, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Trent, D.W. Emergence of epidemic dengue/dengue hemorrhagic fever as a public health problem in the Americas. Infect. Agents Dis. 1993, 2, 383–393. Available online: https://www.paho.org/en/topics/dengue (accessed on 19 May 2023).
- Kouri, G.P.; Guzmán, M.G.; Bravo, J.R.; Triana, C. Dengue Haemorrhagic Fever/Dengue Shock Syndrome: Lessons from the Cuban Epidemic, 1981. Bull. World Health Organ. 1989, 67, 375–380. [Google Scholar]
- L’Azou, M.; Taurel, A.-F.; Flamand, C.; Quénel, P. Recent Epidemiological Trends of Dengue in the French Territories of the Americas (2000–2012): A Systematic Literature Review. PLoS Negl. Trop. Dis. 2014, 8, e3235. [Google Scholar] [CrossRef]
- Matheus, S.; Chappert, J.-L.; Cassadou, S.; Berger, F.; Labeau, B.; Bremand, L.; Winicki, A.; Huc-Anais, P.; Quenel, P.; Dussart, P. Virological Surveillance of Dengue in Saint Martin and Saint Barthélemy, French West Indies, Using Blood Samples on Filter Paper. Am. J. Trop. Med. Hyg. 2012, 86, 159–165. [Google Scholar] [CrossRef]
- Pinheiro, F.; Nelson, M. Re-Emergence of Dengue and Emergence of Dengue Haemorrhagic Fever in the Americas; WHO Regional Office for South-East Asia: New Delhi, India, 1997; Volume 21. [Google Scholar]
- Santé Publique France Points Épidémiologiques Régionaux de La Dengue. Available online: https://www.santepubliquefrance.fr/recherche/#search=&themes=dengue&publications=donn%C3%A9es®ions=Antilles&sort=date (accessed on 31 May 2023).
- Peyrefitte, C.N.; Couissinier-Paris, P.; Mercier-Perennec, V.; Bessaud, M.; Martial, J.; Kenane, N.; Durand, J.-P.A.; Tolou, H.J. Genetic Characterization of Newly Reintroduced Dengue Virus Type 3 in Martinique (French West Indies). J. Clin. Microbiol. 2003, 41, 5195–5198. [Google Scholar] [CrossRef]
- Dussart, P.; Lavergne, A.; Lagathu, G.; Lacoste, V.; Martial, J.; Morvan, J.; Cesaire, R. Reemergence of Dengue Virus Type 4, French Antilles and French Guiana, 2004–2005. Emerg. Infect. Dis. 2006, 12, 1748–1751. [Google Scholar] [CrossRef] [PubMed]
- Callahan, J.D.; Wu, S.-J.L.; Dion-Schultz, A.; Mangold, B.E.; Peruski, L.F.; Watts, D.M.; Porter, K.R.; Murphy, G.R.; Suharyono, W.; King, C.-C.; et al. Development and Evaluation of Serotype- and Group-Specific Fluorogenic Reverse Transcriptase PCR (TaqMan) Assays for Dengue Virus. J. Clin. Microbiol. 2001, 39, 4119–4124. [Google Scholar] [CrossRef] [PubMed]
- Leparc-Goffart, I.; Baragatti, M.; Temmam, S.; Tuiskunen, A.; Moureau, G.; Charrel, R.; de Lamballerie, X. Development and Validation of Real-Time One-Step Reverse Transcription-PCR for the Detection and Typing of Dengue Viruses. J. Clin. Virol. 2009, 45, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. CDC DENV-1-4 RT-PCR Multiplex|CDC. Available online: https://www.cdc.gov/dengue/healthcare-providers/testing/molecular-tests/realtime.html (accessed on 10 July 2023).
- Baronti, C.; Piorkowski, G.; Leparc-Goffart, I.; de Lamballerie, X.; Dubot-Pérès, A. Rapid Next-Generation Sequencing of Dengue, EV-A71 and RSV-A Viruses. J. Virol. Methods 2015, 226, 7–14. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 2005, 21, 98–102. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef] [PubMed]
- BioEdit Version 7.0.0. Available online: https://loschmidt.chemi.muni.cz/bioinf/files/BioEdit_help.pdf (accessed on 7 February 2004).
- Césaire, R.; Cabie, A.; Djossou, F.; Lamaury, I.; Beaucaire, G.; Thomas, L.; Hatchuel, Y.; Najioullah, F.; Yebakima, A.; Cardoso, T.; et al. Aspects Récents de La Dengue Dans Les Départements Français d’Amérique [En Français]. Virologie 2008, 12, 151–157. [Google Scholar] [PubMed]
- Villabona-Arenas, C.J.; Zanotto, P.M. de A. Worldwide Spread of Dengue Virus Type 1. PLoS ONE 2013, 8, e62649. [Google Scholar] [CrossRef] [PubMed]
- Walimbe, A.M.; Lotankar, M.; Cecilia, D.; Cherian, S.S. Global Phylogeography of Dengue Type 1 and 2 Viruses Reveals the Role of India. Infect. Genet. Evol. 2014, 22, 30–39. [Google Scholar] [CrossRef] [PubMed]
- De Bruycker-Nogueira, F.; Mir, D.; dos Santos, F.B.; Bello, G. Evolutionary History and Spatiotemporal Dynamics of DENV-1 Genotype V in the Americas. Infect. Genet. Evol. 2016, 45, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Hamel, R.; Surasombatpattana, P.; Wichit, S.; Dauvé, A.; Donato, C.; Pompon, J.; Vijaykrishna, D.; Liegeois, F.; Vargas, R.M.; Luplertlop, N.; et al. Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand. PLoS ONE 2019, 14, e0221179. [Google Scholar] [CrossRef] [PubMed]
- Calvez, E.; Bounmany, P.; Balière, C.; Somlor, S.; Viengphouthong, S.; Xaybounsou, T.; Keosenhom, S.; Fangkham, K.; Brey, P.T.; Caro, V.; et al. Using Background Sequencing Data to Anticipate DENV-1 Circulation in the Lao PDR. Microorganisms 2021, 9, 2263. [Google Scholar] [CrossRef]
- Dubot-Pérès, A.; Vongphrachanh, P.; Denny, J.; Phetsouvanh, R.; Linthavong, S.; Sengkeopraseuth, B.; Khasing, A.; Xaythideth, V.; Moore, C.E.; Vongsouvath, M.; et al. An Epidemic of Dengue-1 in a Remote Village in Rural Laos. PLoS Neglected Trop. Dis. 2013, 7, e2360. [Google Scholar] [CrossRef]
- Carrillo-Hernandez, M.Y.; Ruiz-Saenz, J.; Jaimes-Villamizar, L.; Robledo-Restrepo, S.M.; Martinez-Gutierrez, M. Phylogenetic and evolutionary analysis of dengue virus serotypes circulating at the Colombian–Venezuelan border during 2015–2016 and 2018–2019. PLoS ONE 2021, 16, e0252379. [Google Scholar] [CrossRef]
- Gupta, A.K.; Kumar, M. Benchmarking and Assessment of Eight De Novo Genome Assemblers on Viral Next-Generation Sequencing Data, Including the SARS-CoV-2. OMICS J. Integr. Biol. 2022, 26, 372–381. [Google Scholar] [CrossRef]
- Eldigail, M.H.; Abubaker, H.A.; Khalid, F.A.; Abdallah, T.M.; Musa, H.H.; Ahmed, M.E.; Adam, G.K.; Elbashir, M.I.; Aradaib, I.E. Association of Genotype III of Dengue Virus Serotype 3 with Disease Outbreak in Eastern Sudan, 2019. Virol. J. 2020, 17, 118. [Google Scholar] [CrossRef] [PubMed]
- Muthanje, E.M.; Kimita, G.; Nyataya, J.; Njue, W.; Mulili, C.; Mugweru, J.; Mutai, B.; Kituyi, S.N.; Waitumbi, J. March 2019 Dengue Fever Outbreak at the Kenyan South Coast Involving Dengue Virus Serotype 3, Genotypes III and V. PLoS Glob. Public Health 2022, 2, e0000122. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.E.; Msafiri, F.; Affara, M.; Gehre, F.; Moremi, N.; Mghamba, J.; Misinzo, G.; Thye, T.; Gatei, W.; Whistler, T.; et al. Molecular Characterization and Phylogenetic Analysis of Dengue Fever Viruses in Three Outbreaks in Tanzania Between 2017 and 2019. PLoS Neglected Trop. Dis. 2023, 17, e0011289. [Google Scholar] [CrossRef] [PubMed]
- Ayolabi, C.I.; Olusola, B.A.; Ibemgbo, S.A.; Okonkwo, G.O. Detection of Dengue Viruses among Febrile Patients in Lagos, Nigeria and Phylogenetics of Circulating Dengue Serotypes in Africa. Infect. Genet. Evol. 2019, 75, 103947. [Google Scholar] [CrossRef]
- Abe, H.; Ushijima, Y.; Loembe, M.M.; Bikangui, R.; Nguema-Ondo, G.; Mpingabo, P.I.; Zadeh, V.R.; Pemba, C.M.; Kurosaki, Y.; Igasaki, Y.; et al. Re-Emergence of Dengue Virus Serotype 3 Infections in Gabon in 2016–2017, and Evidence for the Risk of Repeated Dengue Virus Infections. Int. J. Infect. Dis. 2020, 91, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- Lao, M.; Caro, V.; Thiberge, J.-M.; Bounmany, P.; Vongpayloth, K.; Buchy, P.; Duong, V.; Vanhlasy, C.; Hospied, J.-M.; Thongsna, M.; et al. Co-Circulation of Dengue Virus Type 3 Genotypes in Vientiane Capital, Lao PDR. PLoS ONE 2014, 9, e115569. [Google Scholar] [CrossRef]
- Sang, S.; Yue, Y.; Wang, Y.; Zhang, X. The Epidemiology and Evolutionary Dynamics of Massive Dengue Outbreak in China, 2019. Front. Microbiol. 2023, 14, 1156176. [Google Scholar] [CrossRef]
- Parveen, N.; Islam, A.; Tazeen, A.; Hisamuddin, M.; Abdullah, M.; Naqvi, I.H.; Faizan, M.I.; Gulyani, D.; Ahmed, A.; Parveen, S. Circulation of Single Serotype of Dengue Virus (DENV-3) in New Delhi, India during 2016: A Change in the Epidemiological Trend. J. Infect. Public Health 2019, 12, 49–56. [Google Scholar] [CrossRef]
- INSEE Le Tourisme Aux Antilles. Available online: https://www.insee.fr/fr/statistiques/3651505#tableau-figure2 (accessed on 13 June 2023).
- Leparc-Goffart, I.; Nougairede, A.; Cassadou, S.; Prat, C.; de Lamballerie, X. Chikungunya in the Americas. Lancet 2014, 383, 514. [Google Scholar] [CrossRef]
- Cao-Lormeau, V.-M. Tropical Islands as New Hubs for Emerging Arboviruses. Emerg. Infect. Dis. 2016, 22, 913–915. [Google Scholar] [CrossRef] [PubMed]
- Santé Publique France COVID-19-BVS. Available online: https://www.santepubliquefrance.fr/recherche/#search=Antilles&publications=article|avis|congr%C3%A8s,%20colloque|guides,%20m%C3%A9thodes,%20r%C3%A9f%C3%A9rentiels|magazines,%20revues|rapport,%20synth%C3%A8se|th%C3%A8ses,%20m%C3%A9moires|donn%C3%A9es|BEH|sant%C3%A9%20en%20action®ions=Antilles (accessed on 23 June 2023).
- Brady, O.; Wilder-Smith, A. What Is the Impact of Lockdowns on Dengue? Curr. Infect. Dis. Rep. 2021, 23, 2. [Google Scholar] [CrossRef]
- Yan, G.; Lee, C.K.; Lam, L.T.M.; Yan, B.; Chua, Y.X.; Lim, A.Y.N.; Phang, K.F.; Kew, G.S.; Teng, H.; Ngai, C.H.; et al. Covert COVID-19 and False-Positive Dengue Serology in Singapore. Lancet Infect. Dis. 2020, 20, 536. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Dong, S.; Li, X.; Yi, B.; Hu, H.; Guo, Z.; Lu, J. Effects of COVID-19 Non-Pharmacological Interventions on Dengue Infection: A Systematic Review and Meta-Analysis. Front. Cell Infect. Microbiol. 2022, 12, 892508. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, L. Quantitative Genetics of Aedes Aegypti Vector Competence for Dengue Viruses: Towards a New Paradigm? Trends Parasitol. 2011, 27, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Gloria-Soria, A.; Ayala, D.; Bheecarry, A.; Calderon-Arguedas, O.; Chadee, D.D.; Chiappero, M.; Coetzee, M.; Elahee, K.B.; Fernandez-Salas, I.; Kamal, H.A.; et al. Global Genetic Diversity of Aedes Aegypti. Mol. Ecol. 2016, 25, 5377–5395. [Google Scholar] [CrossRef] [PubMed]
- Viglietta, M.; Bellone, R.; Blisnick, A.A.; Failloux, A.-B. Vector Specificity of Arbovirus Transmission. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Vazeille, M.; Gaborit, P.; Mousson, L.; Girod, R.; Failloux, A.-B. Competitive Advantage of a Dengue 4 Virus When Co-Infecting the Mosquito Aedes Aegypti with a Dengue 1 Virus. BMC Infect. Dis. 2016, 16, 318. [Google Scholar] [CrossRef]




| Sample Identification | Number of Nucleotides | Origin | Year of Collection | Isolation Source | Serotype | Genbank Number |
|---|---|---|---|---|---|---|
| Guadeloupe (IPG5), 2020 | 10,665 | Guadeloupe | July 2020 | Plasma | DENV-1 | OR229954 |
| Guadeloupe (IPG8), 2020 | 10,665 | Guadeloupe | October 2020 | Plasma | DENV-1 | OR229957 |
| Guadeloupe (IPG11), 2020 | 10,665 | Guadeloupe | November 2020 | Plasma | DENV-1 | OR229955 |
| Guadeloupe (IPG12), 2020 | 10,665 | Guadeloupe | November 2020 | Plasma | DENV-1 | OR229956 |
| Guadeloupe (CHUG1), 2020 | 10,667 | Guadeloupe | October 2019 | Plasma | DENV-2 | OR229961 |
| Guadeloupe (CHUG2), 2019 | 10,667 | Guadeloupe | December 2019 | Plasma | DENV-2 | OR229974 |
| Guadeloupe (CHUG3), 2020 | 10,667 | Guadeloupe | January 2020 | Plasma | DENV-2 | OR229965 |
| Guadeloupe (CHUG4), 2020 | 10,667 | Guadeloupe | July 2020 | Plasma | DENV-2 | OR229963 |
| Guadeloupe (CHUG6), 2020 | 10,667 | Guadeloupe | September 2020 | Plasma | DENV-2 | OR229959 |
| Guadeloupe (CHUG8), 2020 | 10,667 | Guadeloupe | October 2020 | Plasma | DENV-2 | OR229962 |
| Guadeloupe (IPG1), 2020 | 10,667 | Guadeloupe | September 2019 | Plasma | DENV-2 | OR229973 |
| Guadeloupe (IPG2), 2020 | 10,667 | Guadeloupe | November 2019 | Plasma | DENV-2 | OR229976 |
| Guadeloupe (IPG6), 2020 | 10,667 | Guadeloupe | July 2020 | Plasma | DENV-2 | OR229958 |
| Guadeloupe (IPG7), 2020 | 10,667 | Guadeloupe | August 2020 | Plasma | DENV-2 | OR229964 |
| Guadeloupe (IPG10), 2020 | 10,667 | Guadeloupe | October 2020 | Plasma | DENV-2 | OR229960 |
| Guadeloupe (IPG14), 2020 | 10,667 | Guadeloupe | November 2020 | Plasma | DENV-2 | OR229975 |
| Martinique (CHUM2), 2020 | 10,667 | Martinique | June 2020 | Plasma | DENV-2 | OR229971 |
| Martinique (CHUM3), 2020 | 10,667 | Martinique | June 2020 | Plasma | DENV-2 | OR229967 |
| Martinique (CHUM6), 2020 | 10,667 | Martinique | June 2020 | Plasma | DENV-2 | OR229970 |
| Martinique (CHUM14), 2020 | 10,667 | Martinique | July 2020 | Plasma | DENV-2 | OR229969 |
| Martinique (CHUM18), 2020 | 10,667 | Martinique | August 2020 | Plasma | DENV-2 | OR229966 |
| Martinique (CHUM20), 2020 | 10,667 | Martinique | August 2020 | Plasma | DENV-2 | OR229972 |
| Martinique (CHUM26), 2020 | 10,667 | Martinique | September 2020 | Plasma | DENV-2 | OR229968 |
| Guadeloupe (IPG16), 2020 | 10,649 | Guadeloupe | December 2020 | Plasma | DENV-3 | OR229980 |
| Guadeloupe (IPG17), 2020 | 10,649 | Guadeloupe | October 2020 | Plasma | DENV-3 | OR229981 |
| Martinique (CHUM11), 2020 | 10,649 | Martinique | July 2020 | Plasma | DENV-3 | OR229984 |
| Martinique (CHUM13), 2020 | 10,649 | Martinique | July 2020 | Plasma | DENV-3 | OR229982 |
| Martinique (CHUM16), 2020 | 10,649 | Martinique | August 2020 | Plasma | DENV-3 | OR229977 |
| Martinique (CHUM17), 2020 | 10,649 | Martinique | August 2020 | Plasma | DENV-3 | OR229979 |
| Martinique (CHUM21), 2020 | 10,649 | Martinique | September 2020 | Plasma | DENV-3 | OR229983 |
| Martinique (CHUM22), 2020 | 10,649 | Martinique | September 2020 | Plasma | DENV-3 | OR229985 |
| Martinique (CHUM24), 2020 | 10,649 | Martinique | September 2020 | Plasma | DENV-3 | OR229978 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia--Van Smévoorde, M.; Piorkowski, G.; Emboulé, L.; Dos Santos, G.; Loraux, C.; Guyomard-Rabenirina, S.; Joannes, M.-O.; Fagour, L.; Najioullah, F.; Cabié, A.; et al. Phylogenetic Investigations of Dengue 2019–2021 Outbreak in Guadeloupe and Martinique Caribbean Islands. Pathogens 2023, 12, 1182. https://doi.org/10.3390/pathogens12091182
Garcia--Van Smévoorde M, Piorkowski G, Emboulé L, Dos Santos G, Loraux C, Guyomard-Rabenirina S, Joannes M-O, Fagour L, Najioullah F, Cabié A, et al. Phylogenetic Investigations of Dengue 2019–2021 Outbreak in Guadeloupe and Martinique Caribbean Islands. Pathogens. 2023; 12(9):1182. https://doi.org/10.3390/pathogens12091182
Chicago/Turabian StyleGarcia--Van Smévoorde, Margot, Géraldine Piorkowski, Loic Emboulé, Georges Dos Santos, Cécile Loraux, Stéphanie Guyomard-Rabenirina, Marie-Odile Joannes, Laurence Fagour, Fatiha Najioullah, André Cabié, and et al. 2023. "Phylogenetic Investigations of Dengue 2019–2021 Outbreak in Guadeloupe and Martinique Caribbean Islands" Pathogens 12, no. 9: 1182. https://doi.org/10.3390/pathogens12091182
APA StyleGarcia--Van Smévoorde, M., Piorkowski, G., Emboulé, L., Dos Santos, G., Loraux, C., Guyomard-Rabenirina, S., Joannes, M. -O., Fagour, L., Najioullah, F., Cabié, A., de Lamballerie, X., Vega-Rúa, A., Césaire, R., & Calvez, E. (2023). Phylogenetic Investigations of Dengue 2019–2021 Outbreak in Guadeloupe and Martinique Caribbean Islands. Pathogens, 12(9), 1182. https://doi.org/10.3390/pathogens12091182