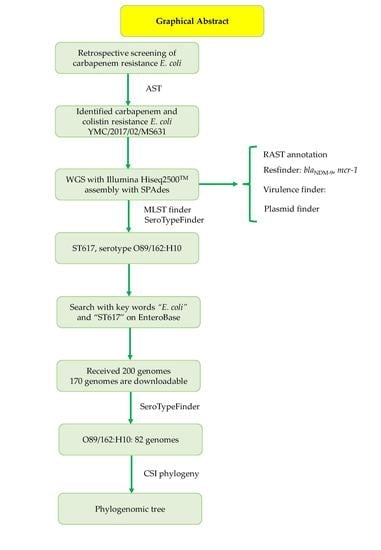
Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea
Abstract
:1. Introduction
2. Case Report
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
Ethics Approval and Consent to Participate
Abbreviations
References
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Zhao, C.; Chen, H.; Liu, J.; Wang, Z.; Wang, Q.; Zhang, Y.; He, W.; Zhang, F.; et al. Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int. J. Antimicrob. Agents 2014, 44, 90–91. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Doi, Y.; Zeng, L.; Lv, L.; Liu, J.-H. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016, 16, 288–289. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Song, F.; Zou, M.; Zhang, Q.; Shan, H. High Incidence of Escherichia coli Strains Coharboring mcr-1 and blaNDM from Chickens. Antimicrob. Agents Chemother. 2017, 61, e02347-16. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-C.; Chuang, Y.-C.; Chen, C.-C.; Tang, H.-J. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents 2017, 49, 517–518. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Kuroda, M.; Suzuki, S.; Mu, J.-J. Emergence of an Escherichia coli strain co-harbouring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J. Glob. Antimicrob. Resist. 2019, 16, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing Twenty-Eighth Informational Supplement M100-S28, 28th ed.; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2018; ISBN 1-562388-39-8. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Cho, Y.J.; Yong, D.; Chun, J. Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J. Bacteriol. 2012, 194, 3742–3743. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Mazzaferri, F.; de Smet, A.M.; Bragantini, D.; Eggimann, P.; Huttner, B.D.; Kuijper, E.J.; Lucet, J.-C.; Mutters, N.T.; Sanguinetti, M.; et al. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin. Microbiol. Infect. 2019, 25, 807–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]

| Parameters | E. coli YMC/2017/ 02/MS631 | Transconjugants | ||
|---|---|---|---|---|
| Selected by Colistin | Selected by Imipenem | E. coli J53 | ||
| E. coli EJ533 | E. coli EJ5331 | |||
| Source | Asymptomatic carrier | - | - | - |
| Isolation site | Rectal swab | - | - | - |
| Resistance genes | blaNDM-9, blaCTX-M-55, blaTEM-1B, aph(3’)IIa, aph(3’)Ib, rmtB, aph(6)-Id, aadA2, oqxA, oqxB, fosA3, mph(A), mdf(A),floR, sul2, tet(A), dfrA12, mcr-1 | mcr-1 | blaNDM-9, aadA2, fosA3, mph(A), dfrA12 | - |
| MLST | 617 | - | - | - |
| Serotype | O89/162:H10 | - | - | - |
| Plasmid replicon type(s) | IncB, IncFII, IncI2, IncN, IncY, IncR, IncX1 | IncI2 | IncB | - |
| Virulence factors | gad, iss | |||
| ompC, ompF | Intact | |||
| MIC (μg/mL, interpretation) | ||||
| Amoxicillin-clavulanic acid | 128, R † | 4, S ‡ | 8, R ‡ | 4, S ‡ |
| Piperacillin | ≥256, R † | N/D | N/D | N/D |
| Piperacillin-tazobactam | ≥256, R † | ≤4, S ‡ | ≥128, R‡ | ≤4, S ‡ |
| Cefotaxime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Ceftazidime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Cefepime | ≥256, R † | ≤1, S ‡ | ≥64, R ‡ | ≤1, S ‡ |
| Cefoxitin | ≥256, R † | 8, S ‡ | 32, R § | ≤1, S ‡ |
| Aztreonam | ≥128, R † | ≤1, S ‡ | ≤1, S ‡ | ≤1, S ‡ |
| Ertapenem | 64, R † | ≤0.5, S ‡ | 4, R ‡ | ≤0.5, S ‡ |
| Meropenem | 16, R † | N/D | N/D | N/D |
| Imipenem | 32, R † | ≤0.25, S ‡ | 8, R ‡ | ≤0.25, S ‡ |
| Ceftazidime-avibactam | ≥256, R † | N/D | N/D | N/D |
| Colistin | 4, R ‡ | 4, R ‡ | ≤0.125, S ‡ | <0.125, S ‡ |
| Amikacin | ≥16, R ‡ | ≤2, S ‡ | ≤2, S ‡ | ≤2, S ‡ |
| Gentamicin | ≥16, R ‡ | ≤1, S ‡ | ≤1, S ‡ | ≤1, S ‡ |
| Ciprofloxacin | ≥4, R ‡ | ≤0.25, S ‡ | ≤0.25, S ‡ | ≤0.25, S ‡ |
| Tigecycline | 0.5, S ‡ | ≤0.5, S ‡ | ≤0.5, S ‡ | ≤0.5, S ‡ |
| Trimethoprim-sulfamethoxazole | 320, R ‡ | ≤20, S ‡ | ≤20, S ‡ | ≤20, S ‡ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, L.P.; Pinto, N.A.; Vu, T.N.; Mai, H.; Pham, A.H.; Lee, H.; Cho, Y.L.; Byun, J.-H.; D’Souza, R.; Yong, D. Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea. Pathogens 2019, 8, 212. https://doi.org/10.3390/pathogens8040212
Nguyen LP, Pinto NA, Vu TN, Mai H, Pham AH, Lee H, Cho YL, Byun J-H, D’Souza R, Yong D. Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea. Pathogens. 2019; 8(4):212. https://doi.org/10.3390/pathogens8040212
Chicago/Turabian StyleNguyen, Le Phuong, Naina Adren Pinto, Thao Nguyen Vu, Hung Mai, An HT Pham, Hyunsook Lee, Young Lag Cho, Jung-Hyun Byun, Roshan D’Souza, and Dongeun Yong. 2019. "Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea" Pathogens 8, no. 4: 212. https://doi.org/10.3390/pathogens8040212
APA StyleNguyen, L. P., Pinto, N. A., Vu, T. N., Mai, H., Pham, A. H., Lee, H., Cho, Y. L., Byun, J. -H., D’Souza, R., & Yong, D. (2019). Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea. Pathogens, 8(4), 212. https://doi.org/10.3390/pathogens8040212