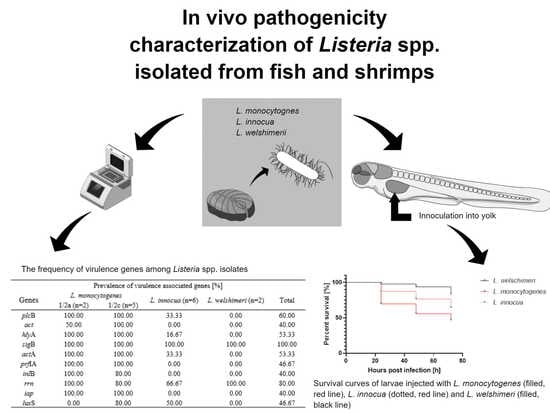
Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacteria
4.3. Pathogenicity Assay Using Zebrafish Larvae
4.4. PCR Assay
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6273–6282. [Google Scholar] [CrossRef] [Green Version]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Guillet, C.; Join-Lambert, O.; Le Monnier, A.; Leclercq, A.; Mechaï, F.; Mamzer-Bruneel, M.F.; Bielecka, M.K.; Scortti, M.; Disson, O.; Berche, P.; et al. Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 2010, 16, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Sauders, B.D.; Overdevest, J.; Fortes, E.; Windham, K.; Schukken, Y.; Lembo, A.; Wiedmann, M. Diversity of Listeria species in urban and natural environments. Appl. Environ. Microbiol. 2012, 78, 4420–4433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snapir, Y.M.; Vaisbein, E.; Nassar, F. Low virulence but potentially fatal outcome-Listeria ivanovii. Eur. J. Intern. Med. 2006, 17, 286–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Molecular Approaches to the Identification of Pathogenic and Nonpathogenic Listeriae. Microbiol. Insights 2013, 6, MBI.S10880. [Google Scholar] [CrossRef] [Green Version]
- Hallstrom, K.N.; McCormick, B.A. Pathogenicity Islands: Origins, Structure, and Roles in Bacterial Pathogenesis. In Molecular Medical Microbiology, 2nd ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; Volume 1–3, pp. 303–314. ISBN 9780123971692. [Google Scholar]
- Papić, B.; Pate, M.; Félix, B.; Kušar, D. Genetic diversity of Listeria monocytogenes strains in ruminant abortion and rhombencephalitis cases in comparison with the natural environment. BMC Microbiol. 2019, 19, 299. [Google Scholar] [CrossRef] [Green Version]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Nørrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16. [Google Scholar] [CrossRef]
- Rocourt, J.; Hof, H.; Schrettenbrunner, A.; Malinverni, R.; Bille, J. Acute purulent Listeria seelingeri meningitis in an immunocompetent adult. Schweiz. Med. Wochenschr. 1986, 116, 248–251. [Google Scholar]
- Poimenidou, S.V.; Dalmasso, M.; Papadimitriou, K.; Fox, E.M.; Skandamis, P.N.; Jordan, K. Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Chan, Y.; Wiedmann, M. Definition of genetically distinct attenuation mechanisms in naturally virulence-attenuated Listeria monocytogenes by comparative cell culture and molecular characterization. Appl. Environ. Microbiol. 2005, 71, 3900–3910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.; Jinneman, K.; Stelma, G.; Smith, B.G.; Lye, D.; Messer, J.; Ulaszek, J.; Evsen, L.; Gendel, S.; Bennett, R.W.; et al. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity island 1 genes. Appl. Environ. Microbiol. 2004, 70, 4256–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, L.Z.; Paixão, R.; de Gobbi, D.D.S.; Raimundo, D.C.; Porfida Ferreira, T.S.; Micke Moreno, A.; Hofer, E.; dos Reis, C.M.F.; Matté, G.R.; Matté, M.H. Phenotypic and genotypic characterization of atypical Listeria monocytogenes and Listeria innocua isolated from swine slaughterhouses and meat markets. Biomed. Res. Int. 2014, 2014, 742032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, K.M.; Samir, A.; Abo-Shama, U.H.; Mohamed, E.H.; Orabi, A.; Zolnikov, T. Determination of virulence and antibiotic resistance pattern of biofilm producing Listeria species isolated from retail raw milk. BMC Microbiol. 2016, 16, 263. [Google Scholar] [CrossRef] [Green Version]
- Wieczorek, K.; Osek, J. Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from fresh and smoked fish in Poland. Food Microbiol. 2017, 64, 164–171. [Google Scholar] [CrossRef]
- Su, X.; Zhang, J.; Shi, W.; Yang, X.; Li, Y.; Pan, H.; Kuang, D.; Xu, X.; Shi, X.; Meng, J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control 2016, 70, 96–102. [Google Scholar] [CrossRef]
- Skowron, K.; Kwiecińska-Piróg, J.; Grudlewska, K.; Świeca, A.; Paluszak, Z.; Bauza-Kaszewska, J.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E. The occurrence, transmission, virulence and antibiotic resistance of Listeria monocytogenes in fish processing plant. Int. J. Food Microbiol. 2018, 282, 71–83. [Google Scholar] [CrossRef]
- Conter, M.; Vergara, A.; Di Ciccio, P.; Zanardi, E.; Ghidini, S.; Ianieri, A. Polymorphism of actA gene is not related to in vitro virulence of Listeria monocytogenes. Int. J. Food Microbiol. 2010, 137, 100–105. [Google Scholar] [CrossRef]
- Stones, D.H.; Fehr, A.G.J.; Thompson, L.; Rocha, J.; Perez-Soto, N.; Madhavan, V.T.P.; Voelz, K.; Krachler, A.M. Zebrafish (Danio rerio) as a Vertebrate Model Host to Study Colonization, Pathogenesis, and Transmission of Foodborne Escherichia coli O157. mSphere 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.J.; Hu, B. Establishment of Multi-Site Infection Model in Zebrafish Larvae for Studying Staphylococcus aureus Infectious Disease. J. Genet. Genom. 2012, 39, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Lamy, B.; Withey, J.H.; Renshaw, S.; Saraceni, P.R.; Romero, A.; Figueras, A.; Novoa, B. Establishment of Infection Models in Zebrafish Larvae (Danio rerio) to Study the Pathogenesis of Aeromonas hydrophila. Front. Microbiol. 2016, 7, 1219. [Google Scholar] [CrossRef] [Green Version]
- Levraud, J.P.; Disson, O.; Kissa, K.; Bonne, I.; Cossart, P.; Herbomel, P.; Lecuit, M. Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect. Immun. 2009, 77, 3651–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menudier, A.; Rougier, F.P.; Bosgiraud, C. Comparative virulence between different strains of Listeria in zebrafish (Brachydanio rerio) and mice. Pathol. Biol. 1996, 44, 783–789. [Google Scholar] [PubMed]
- Favaro, M.; Sarmati, L.; Sancesario, G.; Fontana, C. First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep. 2014, 1, e003103. [Google Scholar] [CrossRef] [Green Version]
- Perrin, M.; Bemer, M.; Delamare, C. Fatal Case of Listeria innocua Bacteremia. J. Clin. Microbiol. 2003, 41, 5308–5309. [Google Scholar] [CrossRef] [Green Version]
- Kluge, R.; Hof, H. Zur Virulenz von Listeria welshimeri. Zent. Bakteriol. Mikrobiol. Hyg. Ser. A Med. Microbiol. Infect. Dis. Virol. Parasitol. 1986, 262, 403–411. [Google Scholar] [CrossRef]
- Mesureur, J.; Vergunst, A.C. Zebrafish embryos as a model to study bacterial virulence. Methods Mol. Biol. 2014, 1197, 41–66. [Google Scholar] [CrossRef]
- Soosai, D.M. Identification of Genetic Determinants Associated with Biofilm Formation Capacity of Listeria Monocytogenes. Master’s Thesis, Univeristy of Ottawa, Ottawa, ON, USA, 2017. [Google Scholar]





| Prevalence of Virulence-Associated Genes (%) | |||||
|---|---|---|---|---|---|
| Genes | L. monocytogenes | L. innocua (n = 6) | L. welshimeri (n = 2) | Total | |
| 1/2a (n = 2) | 1/2c (n = 5) | ||||
| plcB | 100.0 | 100.0 | 33.3 | 0.0 | 60.0 |
| actA2 | 50.0 | 100.0 | 0.0 | 0.0 | 40.0 |
| hlyA | 100.0 | 100.0 | 16.7 | 0.0 | 53.3 |
| sigB | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| actA | 100.0 | 100.0 | 33.3 | 0.0 | 60.0 |
| prfA | 100.0 | 100.0 | 0.0 | 0.0 | 46.7 |
| inlB | 100.0 | 80.0 | 0.0 | 0.0 | 40.0 |
| rrn | 100.0 | 80.0 | 66.7 | 100.0 | 80.0 |
| iap | 100.0 | 100.0 | 0.0 | 0.0 | 40.0 |
| luxS | 0.0 | 80.0 | 50.0 | 0.0 | 46.7 |
| No. | Strain | Species | Serotype | Source |
|---|---|---|---|---|
| 1 | LW104 | L welshimeri | nd. | Salmo salar |
| 2 | LW105 | L welshimeri | nd. | Salmo salar |
| 3 | LI141 | L. innocua | nd. | Oncorhynchus mykiss |
| 4 | LI125 | L. innocua | nd. | Oncorhynchus mykiss |
| 5 | LI113 | L. innocua | nd. | Salmo salar |
| 6 | LI112 | L. innocua | nd. | Salmo salar |
| 7 | LI110 | L. innocua | nd. | Salmo salar |
| 8 | LI103 | L. innocua | nd. | Salmo salar |
| 9 | LM101 | L. monocytogenes | 1/2c | Salmo salar |
| 10 | LM109 | L. monocytogenes | 1/2a | Salmo salar |
| 11 | LM126 | L. monocytogenes | 1/2a | Oncorhynchus mykiss |
| 12 | LM127 | L. monocytogenes | 1/2c | Salmo salar |
| 13 | LM111 | L. monocytogenes | 1/2c | Salmo salar |
| 14 | LM106 | L. monocytogenes | 1/2c | Penaeus monodon |
| 15 | LM107 | L. monocytogenes | 1/2c | Salmo salar |
| No. | Primers | Sequence (5′–3′) | Size (bp) | Target | |
|---|---|---|---|---|---|
| 1 | luxS | F: | ATGGCAGAAAAAATGAATGTAGAAA | 500 | luxS |
| R: | TTATTCACCAAACACATTTTTCCA | ||||
| actA2 | F: | GACGAAAATCCCGAAGTGAA | 385 | actA2 | |
| R: | CTAGCGAAGGTGCTGTTTCC | ||||
| prfA | F: | GATACAGAAACATCGGTTGGC | 280 | prfA | |
| R: | GTGTAATCTTGATGCCATCAGG | ||||
| inlB | F: | AAAGCACGATTTCATGGGAG | 148 | inlB | |
| R: | ACATAGCCTTGTTTGGTCGG | ||||
| rrn | F: | CAG CAG CCG CGG TAA TAC | 938 | rrn | |
| R: | CTC CAT AAA GGT GAC CCT | ||||
| iap | F: | CAAACTGCTAACACAGCTACT | 660 | iap | |
| R: | TTATACGCGACCGAAGCCAAC | ||||
| 2 | sigB | F: | TCATCGGTGTCACGGAAGAA | 310 | sigB |
| R: | TGACGTTGGATTCTAGACAC | ||||
| 3 | plcB | F: | CTGCTTGAGCGTTCATGTCTCATCCCCC | 1150 | plcB |
| R: | ATGGGTTTCACTCTCCTTCTAC | ||||
| actA | F: | CGCCGCGGAAATTAAAAAAAGA | 839 | actA | |
| R: | ACGAAGGAACCGGGCTGCTAG | ||||
| hlyA | F: | GCAGTTGCAAGCGCTTGGAGTGAA | 456 | hlyA | |
| R: | GCAACGTATCCTCCAGAGTGATCG | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakrzewski, A.J.; Chajęcka-Wierzchowska, W.; Zadernowska, A.; Podlasz, P. Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens 2020, 9, 1028. https://doi.org/10.3390/pathogens9121028
Zakrzewski AJ, Chajęcka-Wierzchowska W, Zadernowska A, Podlasz P. Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens. 2020; 9(12):1028. https://doi.org/10.3390/pathogens9121028
Chicago/Turabian StyleZakrzewski, Arkadiusz Józef, Wioleta Chajęcka-Wierzchowska, Anna Zadernowska, and Piotr Podlasz. 2020. "Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study" Pathogens 9, no. 12: 1028. https://doi.org/10.3390/pathogens9121028
APA StyleZakrzewski, A. J., Chajęcka-Wierzchowska, W., Zadernowska, A., & Podlasz, P. (2020). Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens, 9(12), 1028. https://doi.org/10.3390/pathogens9121028