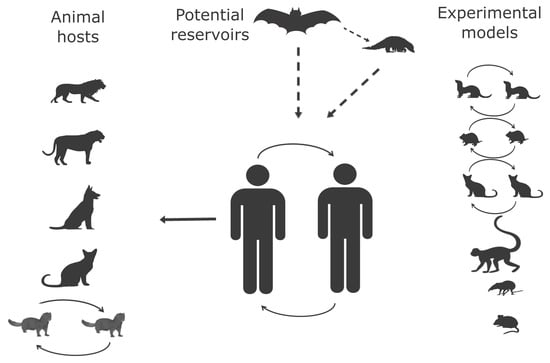
Evidence for SARS-CoV-2 Infection of Animal Hosts
Abstract
:1. Coronaviruses
1.1. Classification of Coronaviruses
1.1.1. Animal Coronaviruses
1.1.2. Human Coronaviruses (HCoVs)
1.2. Coronavirus Structure and Genome Organization
1.3. Genetic Evolution of Coronaviruses
2. SARS-CoV-2
2.1. Animal Hosts
2.1.1. Origin of SARS-CoV-2 and Wild-Animal Reservoir
2.1.2. Natural Infection in Animals
Dogs
Cats
Tigers
Lions
Minks
Other Animals
2.1.3. Experimental Animal Hosts
Rhesus Macaques
Ferrets
Mice
Hamsters
Dogs
Cats
Pigs
Tree Shrew
Bats
Poultry
3. Summary and Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- ICTV. Coronaviridae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae (accessed on 22 June 2020).
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milek, J.; Blicharz-Domanska, K. Coronaviruses in avian species—Review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saif, L.J. Animal coronaviruses: What can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 2004, 23, 643–660. [Google Scholar] [CrossRef]
- Jackwood, M.W. Review of infectious bronchitis virus around the world. Avian. Dis. 2012, 56, 634–641. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, A.N.; Wang, Q.; Jung, K.; Langel, S.N.; Malik, Y.S.; Saif, L.J. Porcine coronaviruses. Emerg. Transbound. Anim. Viruses 2020, 79–110. [Google Scholar] [CrossRef] [Green Version]
- Mora-Diaz, J.C.; Pineyro, P.E.; Houston, E.; Zimmerman, J.; Gimenez-Lirola, L.G. Porcine hemagglutinating encephalomyelitis virus: A review. Front. Vet. Sci. 2019, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Moon, H.; Kang, B. Porcine epidemic diarrhea: A review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015, 4, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Saif, L.J. Bovine respiratory coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef]
- Licitra, B.N.; Duhamel, G.E.; Whittaker, G.R. Canine enteric coronaviruses: Emerging viral pathogens with distinct recombinant spike proteins. Viruses 2014, 6, 3363–3376. [Google Scholar] [CrossRef]
- Erles, K.; Brownlie, J. Canine respiratory coronavirus: An emerging pathogen in the canine infectious respiratory disease complex. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 815–825. [Google Scholar] [CrossRef]
- Olsen, C.W. A review of feline infectious peritonitis virus: Molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993, 36, 1–37. [Google Scholar] [CrossRef]
- Haring, J.; Perlman, S. Mouse hepatitis virus. Curr. Opin. Microbiol. 2001, 4, 462–466. [Google Scholar] [CrossRef]
- Heikkinen, T.; Järvinen, A. The common cold. Lancet 2003, 361, 51–59. [Google Scholar] [CrossRef]
- Makela, M.J.; Puhakka, T.; Ruuskanen, O.; Leinonen, M.; Saikku, P.; Kimpimaki, M.; Blomqvist, S.; Hyypia, T.; Arstila, P. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998, 36, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Gagneur, A.; Sizun, J.; Vallet, S.; Legr, M.C.; Picard, B.; Talbot, P.J. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: A prospective study. J. Hosp. Infect. 2002, 51, 59–64. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Sure, K.; Ihorst, G.; Stang, A.; Pyrc, K.; Jebbink, M.F.; Petersen, G.; Forster, J.; Berkhout, B.; Uberla, K. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005, 2, e240. [Google Scholar] [CrossRef]
- Pyrc, K.; Berkhout, B.; van der Hoek, L. The novel human coronaviruses NL63 and HKU1. J. Virol. 2007, 81, 3051–3057. [Google Scholar] [CrossRef] [Green Version]
- Vijgen, L.; Keyaerts, E.; Moes, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005, 79, 1595–1604. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Sun, K.; Gu, L.; Ma, L.; Duan, Y. Atlas of ACE2 gene expression in mammals reveals novel insights in transmisson of SARS-Cov-2. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013, 100, 246–254. [Google Scholar] [CrossRef]
- Li, B.X.; Ge, J.W.; Li, Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 365, 166–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef] [Green Version]
- Benbacer, L.; Kut, E.; Besnardeau, L.; Laude, H.; Delmas, B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997, 71, 734–737. [Google Scholar] [CrossRef] [Green Version]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669–8674. [Google Scholar] [CrossRef] [Green Version]
- Delmas, B.; Gelfi, J.; L’Haridon, R.; Vogel, L.K.; Sjostrom, H.; Noren, O.; Laude, H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 1992, 357, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Schultze, B.; Krempl, C.; Ballesteros, M.L.; Shaw, L.; Schauer, R.; Enjuanes, L.; Herrler, G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996, 70, 5634–5637. [Google Scholar] [CrossRef] [Green Version]
- Krempl, C.; Schultze, B.; Herrler, G. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 1995, 380, 371–374. [Google Scholar]
- Schultze, B.; Herrler, G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J. Gen. Virol. 1992, 73 Pt 4, 901–906. [Google Scholar] [CrossRef]
- Szczepanski, A.; Owczarek, K.; Bzowska, M.; Gula, K.; Drebot, I.; Ochman, M.; Maksym, B.; Rajfur, Z.; Mitchell, J.A.; Pyrc, K. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: Receptors and attachment factors. Viruses 2019, 11, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; He, B.; Yang, J.; Ye, F.; Lin, S.; Yang, F.; Chen, Z.; Chen, Z.; Cao, Y.; Lu, G. Crystal structure of the S1 subunit N-terminal domain from DcCoV UAE-HKU23 spike protein. Virology 2019, 535, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B.; Wahn, K.; Klenk, H.D.; Herrler, G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 1991, 180, 221–228. [Google Scholar] [CrossRef]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.K.; Jiang, G.S.; Holmes, K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 1991, 88, 5533–5536. [Google Scholar] [CrossRef] [Green Version]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Schultze, B.; Enjuanes, L.; Cavanagh, D.; Herrler, G. N-acetylneuraminic acid plays a critical role for the haemagglutinating activity of avian infectious bronchitis virus and porcine transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 1993, 342, 305–310. [Google Scholar]
- Ambepitiya Wickramasinghe, I.N.; de Vries, R.P.; Weerts, E.A.; van Beurden, S.J.; Peng, W.; McBride, R.; Ducatez, M.; Guy, J.; Brown, P.; Eterradossi, N.; et al. Novel receptor specificity of avian gammacoronaviruses that cause enteritis. J. Virol. 2015, 89, 8783–8792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [PubMed] [Green Version]
- Zeng, Q.H.; Langereis, M.A.; van Vliet, A.L.W.; Huizinga, E.G.; de Groot, R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 9065–9069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escors, D.; Ortego, J.; Laude, H.; Enjuanes, L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001, 75, 1312–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locker, J.K.; Rose, J.K.; Horzinek, M.C.; Rottier, P.J. Membrane assembly of the triple-spanning coronavirus M protein. Individual transmembrane domains show preferred orientation. J. Biol. Chem. 1992, 267, 21911–21918. [Google Scholar] [PubMed]
- Brierley, I.; Digard, P.; Inglis, S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 1989, 57, 537–547. [Google Scholar] [CrossRef]
- Baranov, P.V.; Henderson, C.M.; Anderson, C.B.; Gesteland, R.F.; Atkins, J.F.; Howard, M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 2005, 332, 498–510. [Google Scholar] [CrossRef] [Green Version]
- Mielech, A.M.; Chen, Y.; Mesecar, A.D.; Baker, S.C. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014, 194, 184–190. [Google Scholar] [CrossRef]
- Ziebuhr, J.; Snijder, E.J.; Gorbalenya, A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen Virol. 2000, 81, 853–879. [Google Scholar] [CrossRef]
- Xu, X.; Liu, Y.; Weiss, S.; Arnold, E.; Sarafianos, S.G.; Ding, J. Molecular model of SARS coronavirus polymerase: Implications for biochemical functions and drug design. Nucleic Acids Res. 2003, 31, 7117–7130. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, K.A.; Thiel, V.; Dobbe, J.C.; van der Meer, Y.; Snijder, E.J.; Ziebuhr, J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004, 78, 5619–5632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckerle, L.D.; Becker, M.M.; Halpin, R.A.; Li, K.; Venter, E.; Lu, X.; Scherbakova, S.; Graham, R.L.; Baric, R.S.; Stockwell, T.B.; et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010, 6, e1000896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.; Guan, Y.; Rozanov, M.; Spaan, W.J.; Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- Zhao, L.; Jha, B.K.; Wu, A.; Elliott, R.; Ziebuhr, J.; Gorbalenya, A.E.; Silverman, R.H.; Weiss, S.R. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 2012, 11, 607–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Kamitani, W.; DeDiego, M.L.; Enjuanes, L.; Matsuura, Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012, 86, 11128–11137. [Google Scholar] [CrossRef] [Green Version]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Suzuki, Y.; Gojobori, T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 2004, 21, 1074–1080. [Google Scholar] [CrossRef]
- Ferron, F.; Subissi, L.; Silveira De Morais, A.T.; Le, N.T.T.; Sevajol, M.; Gluais, L.; Decroly, E.; Vonrhein, C.; Bricogne, G.; Canard, B.; et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. USA 2018, 115, E162–E171. [Google Scholar] [CrossRef] [Green Version]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Baric, R.S. Function of a 5’-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J. Virol. 1995, 69, 7529–7540. [Google Scholar] [CrossRef] [Green Version]
- Cavanagh, D.; Mawditt, K.; Adzhar, A.; Gough, R.E.; Picault, J.P.; Naylor, C.J.; Haydon, D.; Shaw, K.; Britton, P. Does IBV Change Slowly Despite the Capacity of the Spike Protein to Vary Greatly. In Coronaviruses and Arteriviruses; Enjuanes, L., Siddell, S.G., Spaan, W., Eds.; Springer US: Boston, MA, USA, 1998; pp. 729–734. [Google Scholar] [CrossRef]
- Pybus, O.; Rambaut, A.; COG-UK-Consortium. Preliminary analysis of SARS-CoV-2 Importation & Establishment of UK Transmission Lineages. Available online: https://virological.org/t/preliminary-analysis-of-sars-cov-2-importation-establishment-of-uk-transmission-lineages/507/2 (accessed on 22 June 2020).
- Fan, Y.; Zhao, K.; Shi, Z.L.; Zhou, P. Bat coronaviruses in China. Viruses 2019, 11, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackwood, M.W.; Hall, D.; Handel, A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012, 12, 1305–1311. [Google Scholar] [CrossRef]
- Lee, C.W.; Jackwood, M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001, 80, 33–39. [Google Scholar] [CrossRef]
- McKinley, E.T.; Hilt, D.A.; Jackwood, M.W. Avian coronavirus infectious bronchitis attenuated live vaccines undergo selection of subpopulations and mutations following vaccination. Vaccine 2008, 26, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, H.; Wu, X.; Zhong, Y.; Zhang, K.; Zhang, Y.P.; Boerwinkle, E.; Fu, Y.X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004, 4, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, S.; Keck, J.G.; Stohlman, S.A.; Lai, M.M. High-frequency RNA recombination of murine coronaviruses. J. Virol. 1986, 57, 729–737. [Google Scholar] [CrossRef] [Green Version]
- Hon, C.C.; Lam, T.Y.; Shi, Z.L.; Drummond, A.J.; Yip, C.W.; Zeng, F.; Lam, P.Y.; Leung, F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008, 82, 1819–1826. [Google Scholar] [CrossRef] [Green Version]
- Stavrinides, J.; Guttman, D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Fan, B.; Chang, X.; Zhou, J.; Zhao, Y.; Shi, D.; Yu, Z.; He, K.; Li, B. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology 2020, 545, 24–32. [Google Scholar] [CrossRef]
- Lu, S.; Wang, Y.; Chen, Y.; Wu, B.; Qin, K.; Zhao, J.; Lou, Y.; Tan, W. Discovery of a novel canine respiratory coronavirus support genetic recombination among betacoronavirus1. Virus Res. 2017, 237, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Luk, H.K.H.; Wong, A.C.P.; Fan, R.Y.Y.; Lam, C.S.F.; Li, K.S.M.; Ahmed, S.S.; Chow, F.W.N.; Cai, J.P.; Zhu, X.; et al. Identification of a novel Betacoronavirus (Merbecovirus) in Amur Hedgehogs from China. Viruses 2019, 11, 980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, M.M.; Graham, R.L.; Donaldson, E.F.; Rockx, B.; Sims, A.C.; Sheahan, T.; Pickles, R.J.; Corti, D.; Johnston, R.E.; Baric, R.S.; et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 19944–19949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnihothram, S.; Yount, B.L., Jr.; Donaldson, E.F.; Huynh, J.; Menachery, V.D.; Gralinski, L.E.; Graham, R.L.; Becker, M.M.; Tomar, S.; Scobey, T.D.; et al. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. mBio 2014, 5, e00047-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef]
- Wang, N.; Li, S.Y.; Yang, X.L.; Huang, H.M.; Zhang, Y.J.; Guo, H.; Luo, C.M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Menachery, V.D.; Yount, B.L., Jr.; Sims, A.C.; Debbink, K.; Agnihothram, S.S.; Gralinski, L.E.; Graham, R.L.; Scobey, T.; Plante, J.A.; Royal, S.R.; et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA 2016, 113, 3048–3053. [Google Scholar] [CrossRef] [Green Version]
- Keha, A.; Xue, L.; Yan, S.; Yue, H.; Tang, C. Prevalence of a novel bovine coronavirus strain with a recombinant hemagglutinin/esterase gene in dairy calves in China. Transbound. Emerg. Dis. 2019, 66, 1971–1981. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Xu, L.; Ren, M.; Shen, J.; Han, Z.; Sun, J.; Zhao, Y.; Liu, S. Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 2019, 230, 178–186. [Google Scholar] [CrossRef]
- Baric, R.S.; Yount, B.; Hensley, L.; Peel, S.A.; Chen, W. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 1997, 71, 1946–1955. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.T.; Chen, T.C.; Lin, S.Y.; Mase, M.; Murakami, S.; Horimoto, T.; Chen, H.W. Emerging lethal infectious bronchitis coronavirus variants with multiorgan tropism. Transbound. Emerg. Dis. 2020, 67, 884–893. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Shi, Z.; Shu, Y.; Song, J.; Gao, G.F.; Tan, W.; Guo, D. A distinct name is needed for the new coronavirus. Lancet 2020, 395, 949. [Google Scholar] [CrossRef]
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 27 April 2020).
- Cleaveland, S.; Laurenson, M.K.; Taylor, L.H. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 991–999. [Google Scholar] [CrossRef]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef]
- Wang, L.F.; Crameri, G. Emerging zoonotic viral diseases. Rev. Sci. Tech. 2014, 33, 569–581. [Google Scholar] [CrossRef]
- Hayman, D.T. Bats as viral reservoirs. Annu. Rev. Virol. 2016, 3, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Bengis, R.G.; Leighton, F.A.; Fischer, J.R.; Artois, M.; Mörner, T.; Tate, C.M. The role of wildlife in emerging and re-emerging zoonoses. Rev. Sci. Tech. 2004, 23, 497–511. [Google Scholar] [PubMed]
- Parrish, C.R.; Holmes, E.C.; Morens, D.M.; Park, E.-C.; Burke, D.S.; Calisher, C.H.; Laughlin, C.A.; Saif, L.J.; Daszak, P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008, 72, 457–470. [Google Scholar] [CrossRef] [Green Version]
- Woolhouse, M.; Scott, F.; Hudson, Z.; Howey, R.; Chase-Topping, M. Human viruses: Discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2864–2871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childs, J.E. Zoonotic viruses of wildlife: Hither from yon. Arch. Virol. 2004, 1–11. [Google Scholar] [CrossRef]
- Bird, B.H.; Mazet, J.A.K. Detection of emerging zoonotic pathogens: An integrated one health approach. Annu. Rev. Anim. Biosci. 2018, 6, 121–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Hu, B.; Zeng, L.P.; Yang, X.L.; Ge, X.Y.; Zhang, W.; Li, B.; Xie, J.Z.; Shen, X.R.; Zhang, Y.Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Paraskevis, D.; Kostaki, E.G.; Magiorkinis, G.; Panayiotakopoulos, G.; Sourvinos, G.; Tsiodras, S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020, 79, 104212. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.; Castoe, T.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020, 30, 2196–2203.e2193. [Google Scholar] [CrossRef]
- Liu, P.; Chen, W.; Chen, J.P. Viral metagenomics revealed Sendai Virus and coronavirus Infection of Malayan Pangolins (Manis javanica). Viruses 2019, 11, 979. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Cui, J.; Qian, Z.; Wang, Y.; Zhang, H.; Duan, Y.; Wu, X.; Yao, X.; Song, Y.; Li, X.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023. [Google Scholar]
- Wong, M.C.; Javornik Cregeen, S.J.; Ajami, N.J.; Petrosino, J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lam, T.T.; Jia, N.; Zhang, Y.W.; Shum, M.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Pond, S.L.K.; Nekrutenko, A.; Nielsen, R. Testing recombination in the pandemic SARS-CoV-2 strains. Available online: https://virological.org/t/testing-recombination-in-the-pandemic-sars-cov-2-strains/492 (accessed on 27 June 2020).
- OIE. COVID-19 (SARS-COV-2), Hong Kong (SAR - PRC) 2020. Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=33546 (accessed on 22 June 2020).
- AFCD. A Second Dog Positive for COVID-19; AFCD, Ed.; Agriculture, Fisheries and Conservation Department: Hong Kong, 2020. Available online: https://www.info.gov.hk/gia/general/202003/19/P2020031900606.htm (accessed on 22 June 2020).
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef]
- Temmam, S.; Barbarino, A.; Maso, D.; Behillil, S.; Enouf, V.; Huon, C.; Jaraud, A.; Chevallier, L.; Backovic, M.; Pérot, P.; et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibanez, S.; Santibanez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, H.; Huang, K.; Yang, Y.; Hui, X.; Gao, J.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. SARS-CoV-2 neutralizing serum antibodies in cats: A serological investigation. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- OIE. COVID-19 (SARS-COV-2), Hong Kong (SAR - PRC). Available online: https://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?page_refer=MapFullEventReport&reportid=33832 (accessed on 22 May 2020).
- OIE. A Case of a Belgian Cat Positive for Covid-19. Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/COV-19/Belgium_28.03.20.pdf (accessed on 22 June 2020).
- CDC. Confirmation of COVID-19 in Two Pet Cats in New York. Available online: https://www.cdc.gov/media/releases/2020/s0422-covid-19-cats-NYC.html (accessed on 22 June 2020).
- APHIS. USDA Statement on the Confirmation of COVID-19 in a Tiger in New York. Available online: https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/NY-zoo-covid-19 (accessed on 22 June 2020).
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, The Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Gretebeck, L.M.; Subbarao, K. Animal models for SARS and MERS coronaviruses. Curr. Opin. Virol. 2015, 13, 123–129. [Google Scholar] [CrossRef]
- Safronetz, D.; Geisbert, T.W.; Feldmann, H. Animal models for highly pathogenic emerging viruses. Curr. Opin. Virol. 2013, 3, 205–209. [Google Scholar] [CrossRef]
- Carvalho, C.; Gaspar, A.; Knight, A.; Vicente, L. Ethical and scientific pitfalls concerning laboratory research with Non-Human primates, and possible solutions. Animals (Basel) 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glass, W.G.; Subbarao, K.; Murphy, B.; Murphy, P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004, 173, 4030–4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.; Vogel, L.; Guarner, J.; Hayes, N.; Murphy, B.; Zaki, S.; Subbarao, K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005, 79, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Martina, B.E.; Haagmans, B.L.; Kuiken, T.; Fouchier, R.A.; Rimmelzwaan, G.F.; Van Amerongen, G.; Peiris, J.S.; Lim, W.; Osterhaus, A.D. Virology: SARS virus infection of cats and ferrets. Nature 2003, 425, 915. [Google Scholar] [CrossRef]
- Chu, Y.K.; Ali, G.D.; Jia, F.; Li, Q.; Kelvin, D.; Couch, R.C.; Harrod, K.S.; Hutt, J.A.; Cameron, C.; Weiss, S.R.; et al. The SARS-CoV ferret model in an infection-challenge study. Virology 2008, 374, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Cockrell, A.S.; Peck, K.M.; Yount, B.L.; Agnihothram, S.S.; Scobey, T.; Curnes, N.R.; Baric, R.S.; Heise, M.T. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J. Virol. 2014, 88, 5195–5199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, E.; Prescott, J.; Baseler, L.; Bushmaker, T.; Thomas, T.; Lackemeyer, M.G.; Martellaro, C.; Milne-Price, S.; Haddock, E.; Haagmans, B.L.; et al. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS ONE 2013, 8, e69127. [Google Scholar] [CrossRef] [Green Version]
- Raj, V.S.; Smits, S.L.; Provacia, L.B.; van den Brand, J.M.; Wiersma, L.; Ouwendijk, W.J.; Bestebroer, T.M.; Spronken, M.I.; van Amerongen, G.; Rottier, P.J.; et al. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J. Virol. 2014, 88, 1834–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef]
- Shan, C.; Shi, Z.-L.; Yuan, Z.-M.; Yao, Y.-F.; Yang, X.-L.; Zhou, Y.-W.; Wu, J.; Gao, G.; Peng, Y.; Yang, L.; et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in the Rhesus Macaques. Res. Sq. 2020. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Deng, W.; Gao, H.; Xiao, C.; Liu, J.; Xue, J.; Lv, Q.; Liu, J.; Yu, P.; Xu, Y.; et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Rapid development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef]
- Richard, M.; Kok, A.; de Meulder, D.; Bestebroer, T.M.; Lamers, M.M.; Okba, N.M.A.; van Vlissingen, M.F.; Rockx, B.; Haagmans, B.L.; Koopmans, M.P.G.; et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. bioRxiv 2020. [Google Scholar] [CrossRef]
- FLI. Novel Coronavirus SARS-CoV-2: Fruit Bats and Ferrets are Susceptible, Pigs and Chickens are Not. Available online: https://www.fli.de/en/press/press-releases/press-singleview/novel-coronavirus-sars-cov-2-fruit-bats-and-ferrets-are-susceptible-pigs-and-chickens-are-not/ (accessed on 10 May 2020).
- Beer, M. COVID-19: Experimental Infection of Fruit Bats, Ferrets, Pigs, and Chicken with SARS-CoV-2 at Friedrich-Loeffler-Institut. Available online: https://promedmail.org/promed-post/?id=7205881 (accessed on 10 May 2020).
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. Experimental Transmission Studies of SARS-CoV-2 in Fruit Bats, Ferrets, Pigs and Chickens. 2020. Available online: https://ssrn.com/abstract=3578792 (accessed on 10 May 2020).
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Lei, C.; Qian, K.; Li, T.; Zhang, S.; Fu, W.; Ding, M.; Hu, S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Comm. 2020, 11, 2070. [Google Scholar] [CrossRef] [Green Version]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; George, A.S.; Schafer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H.; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir potently inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Boudewijns, R.; Thibaut, H.J.; Kaptein, S.J.F.; Li, R.; Vergote, V.; Seldeslachts, L.; De Keyzer, C.; Sharma, S.; Jansen, S.; Weyenbergh, J.V.; et al. STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sia, S.F.; Yan, L.-M.; Chin, A.W.H.; Fung, K.; Choy, K.-T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.A.P.M.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020. [Google Scholar] [CrossRef]
- Lau, S.Y.; Wang, P.; Mok, B.W.; Zhang, A.J.; Chu, H.; Lee, A.C.; Deng, S.; Chen, P.; Chan, K.H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.-i.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, J.; Kuang, D.; Xu, J.; Yang, M.; Ma, C.; Zhao, S.; Li, J.; Long, H.; Ding, K.; et al. Susceptibility of tree shrew to SARS-CoV-2 infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jones, K.E.; Purvis, A.; MacLarnon, A.; Bininda-Emonds, O.R.; Simmons, N.B. A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol. Rev. Camb. Philos. Soc. 2002, 77, 223–259. [Google Scholar] [CrossRef] [PubMed]
- Amador, L.I.; Moyers Arévalo, R.L.; Almeida, F.C.; Catalano, S.A.; Giannini, N.P. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J. Mamm. Evol. 2018, 25, 37–70. [Google Scholar] [CrossRef]
- Shi, Z.-L.; Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Suarez, D.L.; Pantin-Jackwood, M.J.; Swayne, D.E.; Lee, S.A.; DeBlois, S.M.; Spackman, E. Lack of susceptibility of poultry to SARS-CoV-2 and MERS-CoV. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; Ma, C.; et al. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, B.N.; Feldmann, F.; Schwarz, B.; Meade-White, K.; Porter, D.P.; Schulz, J.; van Doremalen, N.; Leighton, I.; Yinda, C.K.; Pérez-Pérez, L.; et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Qi, F.; Xu, Y.; Li, F.; Liu, P.; Liu, J.; Bao, L.; Deng, W.; Gao, H.; Xiang, Z.; et al. Age-related rhesus macaques models of COVID-19. Anim. Model. Exp. Med. 2020, 3, 93–97. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Bao, L.; Gao, H.; Xiang, Z.; Qu, Y.; Song, Z.; Gong, S.; Liu, J.; Liu, J.; Yu, P.; et al. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in Rhesus macaques. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.F.; Eaton, B.T. Bats, civets and the emergence of SARS. Curr. Top Microbiol. Immunol. 2007, 315, 325–344. [Google Scholar]
- Weingartl, H.M.; Copps, J.; Drebot, M.A.; Marszal, P.; Smith, G.; Gren, J.; Andova, M.; Pasick, J.; Kitching, P.; Czub, M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 2004, 10, 179–184. [Google Scholar] [CrossRef]
- Chen, D.; Sun, J.; Zhu, J.; Ding, X.; Lan, T.; Zhu, L.; Xiang, R.; Ding, P.; Wang, H.; Wang, X.; et al. Single-cell screening of SARS-CoV-2 target cells in pets, livestock, poultry and wildlife. bioRxiv 2020. [Google Scholar] [CrossRef]
- Amanat, F.; Krammer, F. SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [Google Scholar] [CrossRef]
- Anon. Paul Ehrlich Institute: First Clinical Trial of a COVID-19 Vaccine Authorised in Germany. 2020. Available online: https://www.pei.de/EN/newsroom/press-releases/year/2020/08-first-clinical-trial-sars-cov-2-germany.html (accessed on 22 June 2020).
- Banyard, A.C.; McElhinney, L.M.; Johnson, N.; Fooks, A.R. Introduction History of rabies control by vaccination. Rev. Sci. Tech. 2018, 37, 305–322. [Google Scholar] [CrossRef] [PubMed]

| Genus | Subgenus | Species | Receptor | Reference |
|---|---|---|---|---|
| Alphacoronavirus | Pedacovirus | Porcine epidemic diarrhea virus | APN | [24] |
| Duvinacovirus | Human coronavirus 229E | APN | [25] | |
| Setracovirus | Human coronavirus NL63 | ACE2 | [26] | |
| Rhinacovirus | Swine acute diarrhea syndrome coronavirus | NI | ||
| Tegacovirus | Alphacoronavirus 1 | |||
| Canine coronavirus | APN | [27] | ||
| Feline infectious peritonitis virus | APN | [28] | ||
| Porcine transmissible gastroenteritis virus | APN | [29] | ||
| Porcine respiratory coronavirus | APN | [30] | ||
| Betacoronavirus | Embecovirus | Betacoronavirus 1 | ||
| Human coronavirus OC43 | Neu5,9Ac2 | [31] | ||
| Equine coronavirus | NI | |||
| Bovine coronavirus | Neu5,9Ac2/ HLA-I | [32,33] | ||
| Dromedary camel coronavirus HKU23 | Sugar | [34] | ||
| Canine respiratory coronavirus | HLA-I | [33] | ||
| Porcine hemagglutinating encephalomyelitis virus | Neu5,9Ac2 | [35] | ||
| HCoV-HKU1 | Neu5,9Ac2 | [36] | ||
| MHV | CEACAM1a | [37] | ||
| Merbecovirus | MERS-CoV | DPP4 | [38] | |
| Sarbecovirus | SARS-CoV-1 | ACE2 | [39] | |
| SARS-CoV-2 | ACE2 | [21] | ||
| Gammacoronavirus | Igacovirus | Avian infectious bronchitis virus | Neu5Gc | [40] |
| Turkey coronavirus (TCoV) | non-sialylated type 2 poly-LacNAc | [41] | ||
| Delatacoronavirus | Buldecovirus | porcine deltacoronavirus | NI |
| Species | Genome Organization | S1/S2 |
|---|---|---|
| HCoV-229E | 5’UTR-Rep-S-3a-3b-E-M-N-3’UTR | DGSIIAVQPR↓NVSYD |
| HCoV-NL63 | 5’UTR-Rep-S-3-E-M-N-3’UTR | DGSLIPVRPR↓NSSDN |
| HCoV-OC43 | 5’UTR-Rep 2-HE-S-5-E-M-N-3’UTR | VDYSKNRRSR↓GAITT |
| HCoV-HKU1 | 5’UTR-Rep-HE-S-4-E-M-8-N-3’UTR | SSSSSRRKRR↓SISA |
| MERS-CoV | 5’UTR-Rep-S-3-4a-4b-5-E-M-8b-N-3’UTR | PSTLTPRSCR↓SVPG |
| SARS-CoV-1 | 5’UTR-Rep-S-3a-3b-E-M-7a-7b-8a-8b-9b-N-3’UTR | TVSL....LR↓STGQ |
| SARS-CoV-2 | 5’UTR-Rep-S-3a-E-M-6-7a-7b-8-N-3’UTR | TQTNSPRRAR↓SVAS |
| Animal | Age, Route, Dose | Virus | Symptoms | Pathology, Immunology (Major Changes) | Replication | Seroconversion | References |
|---|---|---|---|---|---|---|---|
| Monkey | Macaca mulatta, adults–old, 4.75 × 106 pfu, IT, IN, OC. | Chinese strain | Elevated body temperature (> 38 °C), decreased bodyweight | Lung radiographic abnormalities, severe gross lesions on lung, heart and stomach and inflammation in liver and heart. Transient increase in blood CD4+ T cells, CD8+ T cells, monocytes. Increased cytokine response | RNA in nasal, throat and anal swabs, blood, fecal samples | >4 dpi | [156] |
| Young M. mulatta, ~2.3 × 106 pfu, IT, IN, OC. | Elevated body temperature (> 38 °C), decreased bodyweight | Lung radiographic abnormalities, severe gross lesions on lung, heart and stomach, and inflammation in liver and heart Transient increase in blood CD4+ T cells, CD8+ T cells, monocytes. Increased cytokine response | RNA in nasal, throat and anal swabs, blood, fecal samples | >4 dpi | [156] | ||
| Macaca fascicularis, 4.75 × 106 pfu, IT, IN, OC. | Transient elevated body temperature (> 38 °C), decreased bodyweight | Lung radiographic abnormalities, severe gross lesions on lung, heart and stomach and inflammation in liver and heart Transient increase in blood CD4+ T cells, CD8+ T cells, monocytes. Increased cytokine response | RNA in nasal, throat and anal swabs, blood, fecal samples | >4 dpi | [156] | ||
| Callithrix jacchus, 106 pfu, IN. | No signs | No severe lesions | RNA in nasal, throat and anal swabs, blood | Negative | [156] | ||
| Cynomolgus Macaques | 4–5 yr, IT and IN, 106 TCID50 | BetaCoV/Munich/BavPat1/2020 | No signs | Consolidated pulmonary tissues | RNA in nasal swabs, nasal cavity, trachea, bronchi, lungs, ileum, tracheo-bronchial lymph nodes, tonsils | 14 dpi | [157] |
| 15–20 yr, IT and IN, 106 TCID50 | One animal had serous nasal discharge | Consolidated pulmonary tissues | RNA in nasal swabs, nasal cavity, trachea, bronchi, lungs, ileum, tracheo-bronchial lymph nodes, tonsils | 14dpi | [157] | ||
| Rhesus Macaques | 2.6 × 106 TCID50, IT, IN, OC and OR | nCoV-WA1-2020 | Changes in respiratory pattern, piloerection, reduced appetite, hunched posture, pale appearance and dehydration | Lung infiltrates by radiographs, interstitial pneumonia, pulmonary edema Leukocytosis, neutrophilia, monocytosis lymphopenia, increased cytokine and chemokines | Nose, throat, rectal swabs, lungs, bronchioalveolar lavage, lymph nodes, GIT tissues | >10 dpi | [130] |
| 2.6 × 106 TCID50, IN, OR, OC and IT | nCoV-WA1-2020 | Increased respiratory rate, difficulty breathing | Lung infiltrates by radiographs | [158] | |||
| 3–5yr, 106 TCID50, IT | WH-09/human/2020/CHN | Bodyweight loss, transient inappetence, tachypnea and hunched posture, bilateral ground-glass opacification of the lungs | Mild-to-moderate interstitial pneumonia | Viral RNA was detected in nasal, oral and anal swabs, as well as in the nose, lung, gut, spinal cord, heart, skeletal muscles and bladder | >14 dpi | [132] | |
| 106 TCID50 IT | CN1, Chinese virus | Severe interstitial pneumonia | RNA in pharynx, crissum, lung, anal swabs by day 3–7 dpi | [133] | |||
| 6–12 yr, 7 × 106 TCID50 IT | IVCAS 6.7512- Wuhan strain | Reduced appetite, bodyweight loss | Interstitial pneumonia | Virus was isolated from oropharyngeal swabs, trachea, bronchus, lungs | 14 and 21 dpi | [131] | |
| 3–5 yr, 106 TCID50, IN | BetaCoV/Wuhan/IVDC-HB-01/2020 | Weight loss, asthenia | Radiographic changes (ground-glass opacities), interstitial pneumonia Declined CD3+/CD8+ and CD3+/CD4+ T cells | Viral RNA in nasal, throat and anal swabs and lungs | 14 dpi | [159] | |
| 15 yr, IN, 106 TCID50 | Weight loss, asthenia | Radiographic changes (ground-glass opacities), severe interstitial pneumonia | Viral RNA in nasal, throat and anal swabs and lungs | 14 dpi | [159] | ||
| 3–5 yr, 106 TCID50 IT | WH-09/human/2020/CHN | Weight loss | Radiographic changes, moderate interstitial pneumonia | Viral RNA in anal swab | [160] | ||
| 3–5 yr, 106 TCID50 OC | No signs | Mild interstitial pneumonia, mild lung lesions | High viral RNA in conjunctival swab | [160] | |||
| 3–5 yr, 106 TCID50, IG | No signs | Radiographic lung changes, | No viral load | [160] | |||
| Ferrets | 105 PFU, IN or IT | F13/environment/2020/Wuhan, (F13-E), and SARS-CoV-2/CTan/human/2020/Wuhan (CTan-H) | Fever and inappetence | Severe lymphoplasmacytic perivasculitis and vasculitis, increased numbers of type II pneumocytes, macrophages, and neutrophils, mild peribronchitis | Viral RNA in nasal, throat and anal swabs, nasal turbinate and soft palate | [134] | |
| 12–20 Mo, IN, 105.5 TCID50 | NMC-nCoV02, Isolate from Korean patient 2020 | Reduced activity, elevated body temperature and occasionally cough | Acute bronchiolitis, viral antigens in nasal turbinate, trachea, lungs, and intestine Increased immune cell infiltration in the respiratory tract | RNA in serum nasal washes, saliva, urine, feces, nasal turbinate, trachea, lungs, intestine, kidneys | Yes | [135] | |
| 6 Mo, IN, 6. 105 TCID50/0.5mL | BetaCoV/Munich/BavPat1/2020 | Viral RNA in nasal, throat and rectal swabs for up to 19 dpi | 21 dpi | [136] | |||
| 9–12 Mo, IN,105 TCID50 | hCoV-522 19/Germany/BavPat1/2020 | No signs | Lesions were mostly restricted to the nasal cavity Perivascular lymphocytic infiltration and minimally increased numbers of alveolar macrophages | Viral RNA in nasal washes, rectal swabs, respiratory tract, intestine, muscle, skin, lymph node, adrenal gland and/or brain tissues | > 8 dpi | [139] | |
| Hamsters | 6–10 Wk, IN, 105 pfu/0.1 mL | Hong Kong strain | Tachypnea, weight loss | Diffuse alveolar damage with extensive apoptosis, severe lung hemorrhage, Proliferative phase of tissue repair, airway and intestinal involvement with virus N-protein expression, high lung viral load Spleen and lymphoid atrophy associated with marked cytokine activation | Nasal turbinate, trachea, lung, intestine, salivary glands, heart, liver, spleen, lymph nodes, kidney, brain and blood | 7 and 14 dpi | [147] |
| 4–5 Wk, IN, 8 × 104 TCID50/80µL | First confirmed COVID–19 Patient in Hong Kong | Ruffled hair coat, bodyweight loss | Pneumonia, lung consolidation CD3 positive T lymphocytes in the peri-bronchial region | Nasal turbinate, lung, kidney, duodenum | 14 dpi | [148] | |
| 7–8 Wk, IN, 1.5 × 105 pfu | Hong Kong strain | Bodyweight loss | Extensive alveolar wall destruction, alveolar space hemorrhage and mononuclear cell infiltration in the lungs of | Virus was isolated in the tracheal and lung tissues | [149] | ||
| 6–8 Wk, 2 × 105 TCID50, IN | Belgium/GHB-03021/2020 | Multifocal necrotizing bronchiolitis massive leukocyte infiltration and edema | High viral RNA loads and infectious titers in the lungs | [146] | |||
| Transgenic hamster, 5–12 Wk, 2 × 105 or 2 × 106 TCID50, IN | Belgium/GHB-03021/2020 | Limited infiltration of polymorphonuclear leukocytes | High viral RNA loads and infectious titers in the lungs, blood, spleen, liver and upper and/or lower GIT | [146] | |||
| Mice | BALB/C, 2 × 105 TCID50, IN | Belgium/GHB-03021/2020 | Mild lung pathology upregulation of antiviral effector molecules | Low-level replication in the lungs | [146] | ||
| SCID mice (lacking functional T and B cells), 2 × 105 TCID50, IN | Belgium/GHB-03021/2020 | Mild lung pathology | Low-level replication in the lungs | [146] | |||
| WT, or Il28r-/- C57BL/6 mice, 2 × 105 TCID50, IN | Belgium/GHB-03021/2020 | Mild lung pathology | Low-level replication in the lungs | [146] | |||
| Ifnar1-/- C57BL/6 mice, 2 × 105 TCID50, IN | Belgium/GHB-03021/2020 | Increased levels of intra-alveolar hemorrhage, peribronchiolar inflammation | enhanced replication in the lung on 3 dpi | [146] | |||
| 6–11 Mo, IN, 105 TCID50/50 μL | BetaCoV/Wuhan/IVDC-HB169 01/2020 | WT-HB-01, no signs | No histopathological changes | No virus was detected in the lungs | [144] | ||
| Transgenic mice, 6–11 Mo, IN, 105 TCID50/50 μL | BetaCoV/Wuhan/IVDC-HB169 01/2020 | hACE2 mice, slight bristles, bodyweight loss | Lung discoloration, damaged, swollen, enlarged, pneumonia with accumulation of lymphocytes and monocytes, macrophages, T and B lymphocytes | Viruses were isolated/detected from the lungs | 21 dpi | [144] | |
| Transgenic mice, 17-week, IN, 103 PFU | 2019-nCoV/USA-WA1/2020 | Female C57Bl/6 Ces1c-/-, bodyweight loss | Lung hemorrhages and dysfunction | Viruses were isolated from the lungs | [145] | ||
| Tree Shrew | 6–12 Mo, 2–4 yr, 5–7 yr, IN, 106 PFU | Transient elevated body temperature | Pathological alterations in lungs, intestines, spleen, brain, heart, liver, pancreas | Viral RNA in nasal, throat, anal swabs and/or blood, lungs, pancreas, uterus, | [151] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-CoV-2 Infection of Animal Hosts. Pathogens 2020, 9, 529. https://doi.org/10.3390/pathogens9070529
Abdel-Moneim AS, Abdelwhab EM. Evidence for SARS-CoV-2 Infection of Animal Hosts. Pathogens. 2020; 9(7):529. https://doi.org/10.3390/pathogens9070529
Chicago/Turabian StyleAbdel-Moneim, Ahmed S., and Elsayed M. Abdelwhab. 2020. "Evidence for SARS-CoV-2 Infection of Animal Hosts" Pathogens 9, no. 7: 529. https://doi.org/10.3390/pathogens9070529
APA StyleAbdel-Moneim, A. S., & Abdelwhab, E. M. (2020). Evidence for SARS-CoV-2 Infection of Animal Hosts. Pathogens, 9(7), 529. https://doi.org/10.3390/pathogens9070529