Biotic Factors Influence Microbiota of Nymph Ticks from Vegetation in Sydney, Australia
Abstract
:1. Introduction
2. Results
2.1. Pilot Study Determined gDNA Isolation and V3-V4 16S rRNA Gene Diversity Profiling Assay Optimal Combination for Tick Microbiota Studies
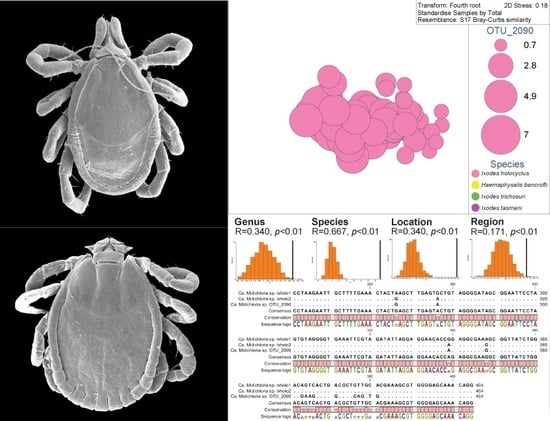
2.2. Ixodes holocyclus, Ixodes trichosuri, Ixodes tasmani and Haemaphysalis bancrofti Nymph Ticks Recovered from Vegetation in Sydney
2.3. Only Adult Ixodes holocyclus Ticks Recovered from Dogs in Sydney Veterinary Clinics
2.4. Pre-Treatment of Adult and Nymph Tick Bacterial Diversity at the 16S rRNA Hypervariable Region V3-V4
2.5. Presence of Candidatus Midichloria spp. in Adult and Nymph Ticks
2.6. Permutation-Based Hypothesis Testing Reveals External Factors Can Influence the Tick’s Microbiota
2.7. Absence of Borrelia spp. in Ixodes holocyclus, Ixodes trichosuri, Ixodes tasmani and Haemaphysalis bancrofti from Sydney
3. Discussion
4. Materials and Methods
4.1. Tick Specimens
4.2. Morphological and Molecular Identity of the Same Ticks Using Two Different Methods
4.3. Morphological Identification and DNA Isolation of Ticks
4.4. Amplification of the Tick Mitochondrially Encoded cox1 Gene
4.5. Tick DNA Sequence Analysis and Phylogeny
4.6. Detection of Borrelia spp. Spirochaete Amplifying a 16S Ribosomal RNA Gene Fragment
4.7. Amplification and Analysis of the Tick Microbial Profile
4.8. Multivariate Statistical Analysis of the Pilot Data for the Single Host Tick Microbiota
4.9. Multivariate Statistical Analysis of the Adult and Nymph Tick Microbiota from Sydney
4.10. Availability of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Statement
References
- Gofton, A.W.; Doggett, S.; Ratchford, A.; Oskam, C.L.; Paparini, A.; Ryan, U.; Irwin, P.; Schneider, B.S. Bacterial profiling reveals novel “Ca. Neoehrlichia”, Ehrlichia, and Anaplasma species in Australian human-biting ticks. PLoS ONE 2015, 10, e0145449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panetta, J.L.; Sima, R.; Calvani, N.E.D.; Hajdušek, O.; Chandra, S.; Panuccio, J.; Šlapeta, J. Reptile-associated Borrelia species in the goanna tick (Bothriocroton undatum) from Sydney, Australia. Parasits Vectors 2017, 10, 616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greay, T.L.; Gofton, A.W.; Paparini, A.; Ryan, U.M.; Oskam, C.L.; Irwin, P.J. Recent insights into the tick microbiome gained through next-generation sequencing. Parasits Vectors 2018, 11, 12. [Google Scholar] [CrossRef]
- Ponnusamy, L.; Gonzalez, A.; Van Treuren, W.; Weiss, S.; Parobek, C.; Juliano, J.; Knight, R.; Roe, R.; Apperson, C.; Meshnick, S. Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum. Appl. Environ. Microbiol. 2014, 80, 354. [Google Scholar] [CrossRef] [Green Version]
- Carpi, G.; Cagnacci, F.; Wittekindt, N.E.; Zhao, F.; Qi, J.; Tomsho, L.P.; Drautz, D.I.; Rizzoli, A.; Schuster, S.C. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS ONE 2011, 6, e25604. [Google Scholar] [CrossRef]
- Ryo, N.; Takashi, A.; Ard, M.N.; Seigo, Y.; Frans, J.; Toshimichi, I.; Chihiro, S. A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks. ISME J. 2013, 7, 1003–1015. [Google Scholar]
- Williams-Newkirk, A.J.; Rowe, L.A.; Mixson-Hayden, T.R.; Dasch, G.A. Characterization of the bacterial communities of life stages of free living lone star ticks (Amblyomma americanum). PLoS ONE 2014, 9, e102130. [Google Scholar] [CrossRef]
- Swei, A.; Kwan, J.Y. Tick microbiome and pathogen acquisition altered by host blood meal. ISME J. 2017, 11, 813–816. [Google Scholar] [CrossRef]
- Kaire, G.H. Isolation of tick paralysis toxin from Ixodes holocyclus. Toxicon 1966, 4, 91–97. [Google Scholar] [CrossRef]
- Ross, I.C. Tick paralysis in the dog caused by nymphs of Ixodes holocyclus. Aust. Vet. J. 1932, 8, 102–104. [Google Scholar] [CrossRef]
- Ross, I.C. An experimental study of tick paralysis in Australia. Parasitology 1926, 18, 410–429. [Google Scholar] [CrossRef]
- Ross, I.C. Tick paralysis: A fatal disease of dogs and other animals in Eastern Australia. J. Council Sci. Ind. Res. 1935, 8, 8–13. [Google Scholar]
- Storer, E.; Sheridan, A.T.; Warren, L.; Wayte, J. Ticks in Australia. Aust. J. Dermatol. 2003, 44, 83–89. [Google Scholar] [CrossRef]
- Brown, A.F.; Hamilton, D.L. Tick bite anaphylaxis in Australia. J. Accid. Emerg. Med. 1998, 15, 111–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gofton, A.W.; Oskam, C.L.; Lo, N.; Beninati, T.; Wei, H.; McCarl, V.; Murray, D.C.; Paparini, A.; Greay, T.L.; Holmes, A.J.; et al. Inhibition of the endosymbiont “Candidatus Midichloria mitochondrii” during 16S rRNA gene profiling reveals potential pathogens in Ixodes ticks from Australia. Parasits Vectors 2015, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Doggett, S.L.; Munro, R.; Ellis, J.; Avery, D.; Hunt, C.; Dickeson, D. Lyme disease: A search for a causative agent in ticks in south-eastern Australia. Epidemiol. Infect. 1994, 112, 375–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, F.H.S. Australian Ticks, 1st ed.; Commonwealth Scientific and Industrial Research Organisation: Melbourne, Australia, 1970; 267p. [Google Scholar]
- Barker, S.C.; Walker, A.R. Ticks of Australia: The species that infest domestic animals and humans. Zootaxa 2014, 3816, 1–144. [Google Scholar] [CrossRef]
- Burgdorfer, W. Discovery of the Lyme disease spirochete and its relation to tick vectors. Yale J. Biol. Med. 1984, 57, 515–520. [Google Scholar]
- Burgdorfer, W.; Barbour, A.G.; Hayes, S.F.; Benach, J.L.; Grunwaldt, E.; Davis, J.P. Lyme disease—A tick-borne spirochetosis? Science 1982, 216, 1317–1319. [Google Scholar] [CrossRef] [Green Version]
- Burgdorfer, W.; Barbour, A.G.; Hayes, S.F.; Peter, O.; Aeschlimann, A. Erythema chronicum migrans—A tickborne spirochetosis. Acta Trop. 1983, 40, 79–83. [Google Scholar]
- Burgdorfer, W.; Lane, R.S.; Barbour, A.G.; Gresbrink, R.A.; Anderson, J.R. The western black-legged tick, Ixodes pacificus: A vector of Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 1985, 34, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Patrican, L.A. Absence of Lyme disease spirochetes in larval progeny of naturally infected Ixodes scapularis (Acari: Ixodidae) fed on dogs. J. Med. Entomol. 1997, 34, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Burgdorfer, W.; Hayes, S.F.; Corwin, D. Pathophysiology of the Lyme disease spirochete, Borrelia burgdorferi, in ixodid ticks. Clin. Infect. Dis. 1989, 11, S1442–S1450. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, M.; Genne, D.; Belli, A.; Maluenda, E.; Sarr, A.; Voordouw, M. The abundance of the Lyme disease pathogen Borrelia afzelii declines over time in the tick vector Ixodes ricinus. Parasits Vectors 2017, 10, 257. [Google Scholar] [CrossRef]
- De Silva, A.M.; Fikrig, E. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am. J. Trop. Med. Hyg. 1995, 53, 397–404. [Google Scholar] [CrossRef]
- Lane, R.S.; Piesman, J.; Burgdorfer, W. Lyme borreliosis: Relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 1991, 36, 587–609. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Kemp, D.H. Identity of Ixodes holocyclus and other paralysis ticks in Australia. Aust. Adv. Vet. Sci. 1979, 1979, 71. [Google Scholar]
- Eisen, R.J.; Eisen, L.; Beard, C.B. County-scale distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the Continental United States. J. Med. Entomol. 2016, 53, 349–386. [Google Scholar] [CrossRef] [Green Version]
- Piesman, J.; Eisen, L. Prevention of Tick-Borne Diseases. Annu. Rev. Entomol. 2008, 53, 323–343. [Google Scholar] [CrossRef]
- Piesman, J.; Oliver, J.R.; Sinsky, R.J. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini). Am. J. Trop. Med. Hyg. 1990, 42, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Mather, T.N.; Nicholson, M.C.; Donnelly, E.F.; Matyas, B.T. Entomologic index for human risk of Lyme disease. Am. J. Epidemiol. 1996, 144, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.M.; Eisen, R.J.; Mead, P.S.; Piesman, J.; Fish, D.; Hoen, A.G.; Barbour, A.G.; Hamer, S.; Diuk-Wasser, M.A. Geographic variation in the relationship between human Lyme disease incidence and density of infected host-seeking Ixodes scapularis nymphs in the Eastern United States. Am. J. Trop. Med. Hyg. 2012, 86, 1062–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stafford, K.C., III; Cartter, M.L.; Magnarelli, L.A.; Ertel, S.-H.; Mshar, P.A. Temporal correlations between tick abundance and prevalence of ticks infected with Borrelia burgdorferi and increasing incidence of Lyme disease. J. Clin. Microbiol. 1998, 36, 1240–1244. [Google Scholar] [CrossRef] [Green Version]
- Loh, S.-M.; Gofton, A.W.; Lo, N.; Gillett, A.; Ryan, U.M.; Irwin, P.J.; Oskam, C.L. Novel Borrelia species detected in echidna ticks, Bothriocroton concolor, in Australia. Parasits Vectors 2016, 9, 339. [Google Scholar] [CrossRef] [Green Version]
- Piesman, J.; Stone, B.F. Vector competence of the Australian paralysis tick, Ixodes holocyclus, for the Lyme disease spirochete Borrelia burgdorferi. Int. J. Parasitol. 1991, 21, 109–111. [Google Scholar] [CrossRef]
- Collignon, P.J.; Lum, G.D.; Robson, J.M.B. Does Lyme disease exist in Australia? Med. J. Aust. 2016, 205, 413–417. [Google Scholar] [CrossRef]
- Irwin, P.J.; Robertson, I.D.; Westman, M.E.; Perkins, M.; Straubinger, R.K. Searching for Lyme borreliosis in Australia: Results of a canine sentinel study. Parasits Vectors 2017, 10, 114. [Google Scholar] [CrossRef] [Green Version]
- Budachetri, K.; Kumar, D.; Crispell, G.; Beck, C.; Dasch, G.; Karim, S. The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the Gulf Coast tick vector. Microbiome 2018, 6, 141. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 1998, 43, 17–37. [Google Scholar] [CrossRef] [Green Version]
- Duron, O.; Morel, O.; Noël, V.; Buysse, M.; Binetruy, F.; Lancelot, R.; Loire, E.; Ménard, C.; Bouchez, O.; Vavre, F.; et al. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr. Biol. 2018, 28, 1896–1902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado-Ferreira, E.; Vizzoni, V.F.; Balsemão-Pires, E.; Moerbeck, L.; Gazeta, G.S.; Piesman, J.; Voloch, C.M.; Soares, C.A. Coxiella symbionts are widespread into hard ticks. Parasitol. Res. 2016, 115, 4691–4699. [Google Scholar] [CrossRef] [PubMed]
- Lalzar, I.; Friedmann, Y.; Gottlieb, Y. Tissue tropism and vertical transmission of Coxiella in Rhipicephalus sanguineus and Rhipicephalus turanicus ticks. Environ. Microbiol. 2014, 16, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, R.; de León, A.A.P.; Dowd, S.E.; Guerrero, F.D.; Bendele, K.G.; Scoles, G.A. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 2011, 11, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sassera, D.; Beninati, T.; Bandi, C.; Bouman, E.A.P.; Sacchi, L.; Fabbi, M.; Lo, N. ‘Candidatus Midichloria mitochondrii’, an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 2006, 56, 2535–2540. [Google Scholar] [CrossRef] [PubMed]
- Ahantarig, A.; Trinachartvanit, W.; Baimai, V.; Grubhoffer, L. Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol. 2013, 58, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Fikrig, E. Tick microbiome: The force within. Trends Parasitol. 2015, 31, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Chicana, B.; Couper, L.I.; Kwan, J.Y.; Tahiraj, E.; Swei, A. Comparative microbiome profiles of sympatric tick species from the Far-Western United States. Insects 2019, 10, 353. [Google Scholar] [CrossRef] [Green Version]
- Thapa, S.; Zhang, Y.; Allen, M.S. Bacterial microbiomes of Ixodes scapularis ticks collected from Massachusetts and Texas, USA. BMC Microbiol. 2019, 19, 138. [Google Scholar] [CrossRef] [Green Version]
- Trout Fryxell, R.T.; DeBruyn, J.M.; Stevenson, B. The microbiome of Ehrlichia-infected and uninfected lone star ticks (Amblyomma americanum). PLoS ONE 2016, 11, e0146651. [Google Scholar]
- Van Treuren, W.; Ponnusamy, L.; Brinkerhoff, R.J.; Gonzalez, A.; Parobek, C.M.; Juliano, J.J.; Andreadis, T.G.; Falco, R.C.; Beati Ziegler, L.; Hathaway, N.; et al. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl. Environ. Microbiol. 2015, 81, 6200–6209. [Google Scholar] [CrossRef] [Green Version]
- Beninati, T.; Riegler, M.; Vilcins, I.M.E.; Sacchi, L.; McFadyen, R.; Krockenberger, M.; Bandi, C.; O’Neill, S.L.; Lo, N. Absence of the symbiont Candidatus Midichloria mitochondrii in the mitochondria of the tick Ixodes holocyclus. FEMS Microbiol. Lett. 2009, 299, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guizzo, M.G.; Neupane, S.; Kucera, M.; Perner, J.; Frantova, H.; Vaz, I.D.S.; De Oliveira, P.L. Poor unstable midgut microbiome of hard ticks contrasts with abundant and stable monospecific microbiome in ovaries. Front. Cell. Infect. Microbiol. 2020, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D. The detection of Rickettsia-like microorganisms within the ovaries of female Ixodes ricinus ticks. Z. Parasitenkd. 1979, 59, 295–298. [Google Scholar] [CrossRef]
- Venere, M.; Fumagalli, M.; Cafiso, A.; Marco, L.; Epis, S.; Plantard, O.; Bardoni, A.; Salvini, R.; Viglio, S.; Bazzocchi, C. Ixodes ricinus and its endosymbiont Midichloria mitochondrii: A comparative proteomic analysis of salivary glands and ovaries. PLoS ONE 2015, 10, e0138842. [Google Scholar] [CrossRef] [Green Version]
- Lo, N.; Beninati, T.; Sassera, D.; Bouman, E.A.P.; Santagati, S.; Gern, L.; Sambri, V.; Masuzawa, T.; Gray, J.S.; Jaenson, T.G.T.; et al. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ. Microbiol. 2006, 8, 1280–1287. [Google Scholar] [CrossRef] [Green Version]
- Klubal, R.; Kopecky, J.; Nesvorna, M.; Sparagano, O.A.; Thomayerova, J.; Hubert, J. Prevalence of pathogenic bacteria in Ixodes ricinus ticks in Central Bohemia. Exp. Appl. Acarol. 2016, 68, 127–137. [Google Scholar] [CrossRef]
- Gofton, A.W.; Doggett, S.; Ratchford, A.; Ryan, U.; Irwin, P. Phylogenetic characterisation of two novel Anaplasmataceae from Australian Ixodes holocyclus ticks: ‘Candidatus Neoehrlichia australis’ and ‘Candidatus Neoehrlichia arcana’. Int. J. Syst. Evol. Microbiol. 2016, 66, 4256–4261. [Google Scholar] [CrossRef]
- Silaghi, C.; Beck, R.; Oteo, J.; Pfeffer, M.; Sprong, H. Neoehrlichiosis: An emerging tick-borne zoonosis caused by Candidatus Neoehrlichia mikurensis. Exp. Appl. Acarol. 2016, 68, 279–297. [Google Scholar] [CrossRef]
- Rynkiewicz, E.C.; Hemmerich, C.; Rusch, D.B.; Fuqua, C.; Clay, K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol. Ecol. 2015, 24, 2566–2579. [Google Scholar] [CrossRef]
- Van Overbeek, L.; Gassner, F.; van der Plas, C.L.; Kastelein, P.; Nunes-da Rocha, U.; Takken, W. Diversity of Ixodes ricinus tick-associated bacterial communities from different forests. FEMS Microbiol. Ecol. 2008, 66, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Sperling, J.L.; Silva-Brandão, K.L.; Brandão, M.M.; Lloyd, V.K.; Dang, S.; Davis, C.S.; Sperling, F.A.H.; Magor, K.E. Comparison of bacterial 16S rRNA variable regions for microbiome surveys of ticks. Ticks Tick Borne Dis. 2017, 8, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Clow, K.M.; Weese, J.S.; Rousseau, J.; Jardine, C.M. Microbiota of field-collected Ixodes scapularis and Dermacentor variabilis from eastern and southern Ontario, Canada. Ticks Tick Borne Dis. 2018, 9, 235–244. [Google Scholar] [CrossRef]
- Hawlena, H.; Rynkiewicz, E.; Toh, E.; Alfred, A.; Durden, L.A.; Hastriter, M.W.; Nelson, D.E.; Rong, R.; Munro, D.; Dong, Q.; et al. The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J. 2013, 7, 221–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, B.E.R.; Sanders, J.G.; Hampton-Marcell, J.; Owens, S.M.; Gilbert, J.A.; Moreau, C.S. DNA extraction protocols cause differences in 16S rRNA amplicon sequencing efficiency but not in community profile composition or structure. MicrobiologyOpen 2014, 3, 910–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishnivetskaya, T.A.; Layton, A.C.; Lau, M.C.; Chauhan, A.; Cheng, K.R.; Meyers, A.J.; Murphy, J.R.; Rogers, A.W.; Saarunya, G.S.; Williams, D.E.; et al. Commercial DNA extraction kits impact observed microbial community composition in permafrost samples. FEMS Microbiol. Ecol. 2014, 87, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kushimo, O.M. The Tick Genus Amblyomma in Africa: Phylogeny and Mutilocus DNA Barcoding. Master’s Thesis, Georgia Southern University, Statesboro, GA, USA, 2013. [Google Scholar]
- Tajima, F.; Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1984, 1, 269–285. [Google Scholar]
- Rzhetsky, A.; Nei, M. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 1992, 9, 945–967. [Google Scholar]
- Kruskal, J.B.; Wish, M. Multidimensional scaling. In Quantitative Applications in the Social Sciences; Uslaner, E.M., Ed.; SAGE Publications Ltd.: Thousand Oaks, CA, USA, 1978; Volume 1, 96p. [Google Scholar]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- NSW Government. Coastal Management Act 2016 No. 20. Available online: https://legislation.nsw.gov.au/#/view/act/2016/20/full (accessed on 3 April 2020).








| S | H’ ± SD | 1-λ’ ± SD | |
|---|---|---|---|
| Nymphs | 2936 | 2.938 ± 0.59 | 0.919 ± 0.07 |
| Adults | 2512 | 2.876 ± 0.82 | 0.882 ± 0.11 |
| Nymphs + Adults | 5448 | 2.912 ± 0.69 | 0.903 ± 0.09 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandra, S.; Šlapeta, J. Biotic Factors Influence Microbiota of Nymph Ticks from Vegetation in Sydney, Australia. Pathogens 2020, 9, 566. https://doi.org/10.3390/pathogens9070566
Chandra S, Šlapeta J. Biotic Factors Influence Microbiota of Nymph Ticks from Vegetation in Sydney, Australia. Pathogens. 2020; 9(7):566. https://doi.org/10.3390/pathogens9070566
Chicago/Turabian StyleChandra, Shona, and Jan Šlapeta. 2020. "Biotic Factors Influence Microbiota of Nymph Ticks from Vegetation in Sydney, Australia" Pathogens 9, no. 7: 566. https://doi.org/10.3390/pathogens9070566
APA StyleChandra, S., & Šlapeta, J. (2020). Biotic Factors Influence Microbiota of Nymph Ticks from Vegetation in Sydney, Australia. Pathogens, 9(7), 566. https://doi.org/10.3390/pathogens9070566