New Molecular Data on Filaria and its Wolbachia from Red Howler Monkeys (Alouatta macconnelli) in French Guiana—A Preliminary Study
Abstract
:1. Introduction
2. Results
2.1. Host Identification
2.2. Molecular Screening for Filarial and Wolbachia DNAs in Howler Monkeys
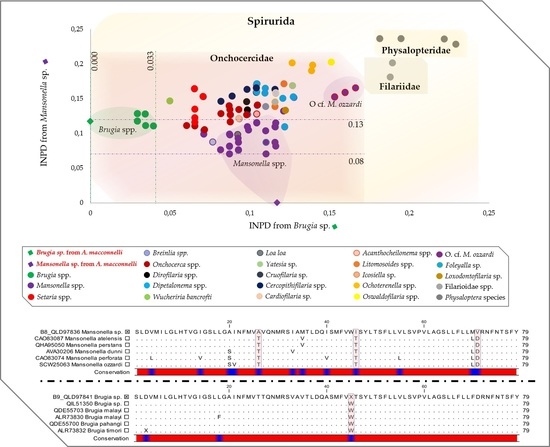
2.3. Molecular Characterization of Filarial Species
3. Discussion
4. Materials and Methods
4.1. Samples and Ethic Statement
4.2. DNA Extraction
4.3. Host Identification
4.4. Molecular Screening for Filaria and Wolbachia
4.5. Molecular Characterization of Filariids and their Associated Wolbachia Using Generic Primers
4.6. Molecular Characterization of Filariids Using Genus Specific PCR Assays
4.6.1. Design of Oligonucleotides
4.6.2. Amplification, Sequencing and Run Protocol
4.6.3. Molecular Screening for Brugia
4.7. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pritt, S.; Cohen, K.; Sedlacek, H. Parasitic Diseases. In The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents, 2nd ed.; American College of Laboratory Animal Medicine; Academic Press: London, UK, 2012; pp. 415–446. ISBN 9780123809209. [Google Scholar]
- Anderson, R.C. Nematode parasites of vertebrates. Their development and transmission; C.A.B. International: Wallingford, UK, 1992; p. 578. [Google Scholar]
- Strait, K.; Else, J.G.; Eberhard, M.L. Parasitic Diseases of Nonhuman Primates. In Nonhuman Primates in Biomedical Research, 2nd ed.; Academic Press Elsevier Inc.: London, UK, 2012; Volume 2, pp. 197–297. ISBN 9780123813664. [Google Scholar]
- Orihel, T.C.; Seibold, H.R. Nematodes of the bowel and tissues. In Pathology of Simian Primates; Fiennes, R.N., Ed.; Karger Publisher: Basel, Switzerland, 1972; pp. 1366–1368. [Google Scholar]
- Gardiner, C.H.; Nold, J.B.; Sanders, J.E. Diagnostic exercise. Edesonfilaria malayensis infection in two cynomolgus monkeys. Lab. Anim. Sci. 1982, 32, 601–602. [Google Scholar]
- Lowenstine, L.J.; Osborn, K.G. Respiratory system diseases of nonhuman primates. In Nonhuman Primates in Biomedical Research, 2nd ed.; Academic Press Elsevier Inc.: London, UK, 2012; Volume 2, pp. 413–481. ISBN 9780123813664. [Google Scholar]
- Bennuru, S.; Nutman, T.B. Lymphangiogenesis and lymphatic remodeling induced by filarial parasites: Implications for pathogenesis. PLoS Pathog. 2009, 5, e1000688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, D.D.; Atkins, C.E. Heartworm biology, treatment, and control. Vet. Clin. Small Anim. Pract. 2009, 39, 1127–1158. [Google Scholar] [CrossRef]
- Fischer, P.; Supali, T.; Maizels, R.M. Lymphatic filariasis and Brugia timori: Prospects for elimination. Trends Parasitol. 2004, 20, 351–355. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Lymphatic Filariasis. Available online: https://www.who.int/news-room/fact-sheets/detail/lymphatic-filariasis (accessed on 12 May 2020).
- Bandi, C.; Anderson, T.J.C.; Genchi, C.; Blaxter, M.L. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. B Biol. Sci. 1998, 265, 2407–2413. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.; Gavotte, L. The bacteria Wolbachia in filariae, a biological Russian dolls’ system: New trends in antifilarial treatments. Parasite 2010, 17, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.J. Wolbachia in the inflammatory pathogenesis of human filariasis. Ann. N. Y. Acad. Sci. 2003, 990, 444–449. [Google Scholar] [CrossRef]
- Dingman, P.; Levy, J.K.; Kramer, L.H.; Johnson, C.M.; Lappin, M.R.; Greiner, E.C.; Courtney, C.H.; Tucker, S.J.; Morchon, R. Association of Wolbachia with heartworm disease in cats and dogs. Vet. Parasitol. 2010, 170, 50–60. [Google Scholar] [CrossRef]
- Bouchery, T.; Lefoulon, E.; Karadjian, G.; Nieguitsila, A.; Martin, C. The symbiotic role of Wolbachia in Onchocercidae and its impact on filariasis. Clin. Microbiol. Infect. 2013, 19, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Hise, A.G.; Gillette-Ferguson, I.; Pearlman, E. The role of endosymbiotic Wolbachia bacteria in filarial disease. Cell. Microbiol. 2004, 6, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Debrah, L.B.; Phillips, R.O.; Pfarr, K.; Klarmann-Schulz, U.; Opoku, V.S.; Nausch, N.; Owusu, W.; Mubarik, Y.; Sander, A.L.; Lämmer, C.; et al. The efficacy of doxycycline treatment on Mansonella perstans infection: An open-label, randomized trial in Ghana. Am. J. Trop. Med. Hyg. 2019, 101, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, W.A.W.; Kamtchum-Tatuene, J.; Mohamed, M.H.; Ramachandran, V.; Ching, S.M.; Lim, S.M.S.; Hashim, H.Z.; Mat, L.N.I.; Hoo, F.K.; Basri, H. Anti-Wolbachia therapy for onchocerciasis & lymphatic filariasis: Current perspectives. Indian J. Med. Res. 2019, 149, 706–714. [Google Scholar]
- McCall, J.W.; Genchi, C.; Kramer, L.; Guerrero, J.; Dzimianski, M.T. Heartworm and Wolbachia: Therapeutic implications. Vet. Parasitol. 2008, 158, 204–214. [Google Scholar] [CrossRef]
- Punkosdy, G.A.; Dennis, V.A.; Lasater, B.L.; Tzertzinis, G.; Foster, J.M.; Lammie, P.J. Detection of serum IgG antibodies specific for Wolbachia surface protein in Rhesus monkeys infected with Brugia malayi. J. Infect. Dis. 2001, 184, 385–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turb, M.E.; Zambon, E.; Zannoni, A.; Russo, S.; Gentilini, F. Detection of Wolbachia DNA in blood for diagnosing filaria-associated syndromes in cats. J. Clin. Microbiol. 2012, 50, 2624–2630. [Google Scholar] [CrossRef] [Green Version]
- Simón, F.; Siles-Lucas, M.; Morchón, R.; González-Miguel, J.; Mellado, I.; Carretón, E.; Montoya-Alonso, J.A. Human and animal dirofilariasis: The emergence of a zoonotic mosaic. Clin. Microbiol. Rev. 2012, 25, 507–544. [Google Scholar] [CrossRef] [Green Version]
- Maia, C.; Altet, L.; Serrano, L.; Cristóvão, J.M.; Tabar, M.D.; Francino, O.; Cardoso, L.; Campino, L.; Roura, X. Molecular detection of Leishmania infantum, filariae and Wolbachia spp. in dogs from southern Portugal. Parasit. Vectors 2016, 9, 170. [Google Scholar] [CrossRef] [Green Version]
- Laidoudi, Y.; Davoust, B.; Varloud, M.; Niang, E.H.A.; Fenollar, F.; Mediannikov, O. Development of a multiplexed qPCRs-based approach for the diagnosis of Dirofilaria immitis, D. repens, Acanthocheilonema reconditum. Parasites Vectors Dev. 2019, 13, 319. [Google Scholar] [CrossRef]
- Laidoudi, Y.; Marié, J.L.; Tahir, D.; Watier-Grillot, S.; Mediannikov, O.; Davoust, B. Detection of canine vector-borne filariasis and their Wolbachia endosymbionts in French Guiana. Microorganisms 2020, 8, 770. [Google Scholar] [CrossRef]
- Ichimori, K.; King, J.D.; Engels, D.; Yajima, A.; Mikhailov, A.; Lammie, P.; Ottesen, E.A. Global programme to eliminate lymphatic filariasis: The processes underlying programme success. PLoS Negl. Trop. Dis. 2014, 8, e3328. [Google Scholar] [CrossRef]
- Webster, J.P.; Molyneux, D.H.; Hotez, P.J.; Fenwick, A. The contribution of mass drug administration to global health: Past, present and future. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130434. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Lymphatic Filariasis: Practical Entomology—A Handbook for National Elimination Programmes; No. WHO/HTM/NTD/PCT/2013.10.; World Health Organization: Geneva, Switzerland, 2013; p. 92. [Google Scholar]
- Hotez, P.J.; Bottazzi, M.E.; Franco-Paredes, C.; Ault, S.K.; Periago, M.R. The neglected tropical diseases of Latin America and the Caribbean: A review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl. Trop. Dis. 2008, 2, e300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, M.C.; Aguilera, X.P.; da Silva Junior, J.B.; Ault, S.K.; Najera, P.; Martinez, J.; Requejo, R.; Nicholls, R.S.; Yadon, Z.; Silva, J.C.; et al. Elimination of neglected diseases in Latin America and the Caribbean: A mapping of selected diseases. PLoS Negl. Trop. Dis. 2011, 5, e964. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organisation and World Health Organisation. Latin American countries gather to work towards the elimination of lymphatic filariasis (elephantiasis). Available online: https://www.paho.org/hq/index.php?option=com_content&view=article&id=8784:2013-paises-latinoamerica-reunen-avanzar-hacia-eliminacion-filariasis-linfatica-elefantiasis&Itemid=40282&lang=en (accessed on 22 June 2020).
- Raccurt, C.P. Mansonella ozzardi and its vectors in the New World: An update with emphasis on the current situation in Haiti. J. Helminthol. 2018, 92, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Ta-Tang, T.H.; Luz, S.L.B.; Merino, F.J.; De Fuentes, I.; López-Vélez, R.; Almeida, T.A.P.; Lanza, M.; Abrahim, C.M.M.; Rubio, J.M. Atypical Mansonella ozzardi microfilariae from an endemic area of Brazilian amazonia. Am. J. Trop. Med. Hyg. 2016, 95, 629–632. [Google Scholar] [CrossRef] [Green Version]
- Tavares da Silva, L.B.; Crainey, J.L.; Ribeiro da Silva, T.R.; Suwa, U.F.; Vicente, A.C.P.; Fernandes de Medeiros, J.; Pessoa, F.A.C.; Luz, S.L.B. Molecular verification of new world Mansonella perstans parasitemias. Emerg. Infect. Dis. 2017, 23, 545–547. [Google Scholar] [CrossRef] [Green Version]
- Orihel, T.C. Brugia guyanensis sp. n. (Nematoda: Filarioidea) from the Coatimundi (Nasua nasua vittata) in British Guiana. J. Parasitol. 1964, 50, 115–118. [Google Scholar] [CrossRef]
- Moraes, M.F.D.; da Silva, M.X.; Magalhães-Matos, P.C.; de Albuquerque, A.C.A.; Tebaldi, J.H.; Mathias, L.A.; Lux Hoppe, E.G. Filarial nematodes with zoonotic potential in ring-tailed coatis (Nasua nasua Linnaeus, 1766, Carnivora: Procyonidae) and domestic dogs from Iguaçu National Park, Brazil. Vet. Parasitol. Reg. Stud. Rep. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- De Argôlo, E.G.G.; Reis, T.; Fontes, D.A.T.; Gonçalves, E.C.; Giese, E.G.; de Vasconcelos Melo, F.T.; dos Santos, J.N.; Furtado, A.P. Canine filariasis in the Amazon: Species diversity and epidemiology of these emergent and neglected zoonoses. PLoS ONE 2018, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, J.F.; Redford, K.H. Alouatta. In Mammals of the Neotropics. The Central Neotropics: Ecuador, Perú, Bolivia, Brazil; University of Chicago Press: Chicago, IL, USA, 1999; Volume 3, pp. 265–266. [Google Scholar]
- Stuart, M.; Pendergast, V.; Rumfelt, S.; Pierberg, S.; Greenspan, L.; Glander, K.; Clarke, M. Parasites of wild howlers (Alouatta spp.). Int. J. Primatol. 1998, 19, 493–512. [Google Scholar] [CrossRef]
- Cortés-Ortiz, L.; Bermingham, E.; Rico, C.; Rodríguez-Luna, E.; Sampaio, I.; Ruiz-García, M. Molecular systematics and biogeography of the Neotropical monkey genus, Alouatta. Mol. Phylogenet. Evol. 2003, 26, 64–81. [Google Scholar] [CrossRef]
- Groves, C. Primate taxonomy. In The International Encyclopedia of Biological Anthropology; John Wiley & Sons: Hoboken, NJ, USA, 2018; Volume 1. [Google Scholar] [CrossRef]
- Stevenson, P.R. Potential determinants of the abundance of woolly monkeys in neotropical forests. In The Woolly Monkey: Behavior, Ecology, Systematics, and Captive Research; Pablo, R., Ed.; Springer: New York, NY, USA, 2014; ISBN 9781493906970. [Google Scholar]
- Medkour, H.; Davoust, B.; Levasseur, A.; Mediannikov, O. Molecular evidence of Leishmania infantum and Leishmania guyanensis in red howler monkey (Alouatta seniculus) from French Guiana. Vector Borne Zoonotic Dis. 2019, 19, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Bain, O.; Mutafchiev, Y.; Junker, K.; Guerrero, R.; Martin, C.; Lefoulon, E.; Uni, S. Review of the genus Mansonella Faust, 1929 sensu lato (Nematoda: Onchocercidae), with descriptions of a new subgenus and a new subspecies. Zootaxa 2015, 3918, 151–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crainey, J.L.; Marín, M.A.; Da Silva, T.R.R.; De Medeiros, J.F.; Pessoa, F.A.C.; Santos, Y.V.; Vicente, A.C.P.; Luz, S.L.B. Mansonella ozzardi mitogenome and pseudogene characterisation provides new perspectives on filarial parasite systematics and CO-1 barcoding. Sci. Rep. 2018, 8, 6158. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-García, M.; Cerón, Á.; Sánchez-Castillo, S.; Rueda-Zozaya, P.; Pinedo-Castro, M.; Gutierrez-Espeleta, G.; Shostell, J.M. Phylogeography of the mantled howler monkey (Alouatta palliata; Atelidae, Primates) across its geographical range by means of mitochondrial genetic analyses and new insights about the phylogeny of Alouatta. Folia Primatol. 2018, 88, 421–454. [Google Scholar] [CrossRef]
- De Thoisy, B.; Michel, J.C.; Vogel, I.; Vié, J.C. A survey of hemoparasite infections in free-ranging mammals and reptiles in French Guiana. J. Parasitol. 2000, 86, 1035–1040. [Google Scholar] [CrossRef]
- Klein, A.; Strube, C.; Radespiel, U.; Springer, A.; Zimmermann, E. Differences in infection patterns of vector-borne blood-stage parasites of sympatric Malagasy primate species (Microcebus murinus, M. ravelobensis). Int. J. Parasitol. Parasites Wildl. 2019, 10, 59–70. [Google Scholar] [CrossRef]
- Lainson, R.; Shaw, J.J.; Fraiha, H.; Miles, M.A.; Draper, C.C. Chagas’s disease in the amazon basin: I. Trypanosoma cruzi infections in silvatic mammals, triatomine bugs and man in the state of North Brazil. Trans. R. Soc. Trop. Med. Hyg. 1979, 73, 193–204. [Google Scholar] [CrossRef]
- Petit, G.; Bain, O.; Roussilhon, C. Deux nouvelles filaires chez un singe, Saimiri sciureus, au Guyana. Ann. Parasitol. Hum. Comp. 1985, 60, 65–81. [Google Scholar] [CrossRef] [Green Version]
- De León, G.P.-P.; Nadler, S.A. What we don’t recognize can hurt us: A plea for awareness about cryptic species. J. Parasitol. 2010, 96, 453–464. [Google Scholar] [CrossRef]
- De Ley, P.; Tandingan De Ley, I.; Morris, K.; Abebe, E.; Mundo-Ocampo, M.; Yoder, M.; Heras, J.; Waumann, D.; Rocha-Olivares, A.; Burr, A.H.J.; et al. An integrated approach to fast and informative morphological vouchering of nematodes for applications in molecular barcoding. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1945–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Meldal, B.H.M.; Debenham, N.J.; De Ley, P.; De Ley, I.T.; Vanfleteren, J.R.; Vierstraete, A.R.; Bert, W.; Borgonie, G.; Moens, T.; Tyler, P.A.; et al. An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol. Phylogenet. Evol. 2007, 42, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Nuchprayoon, S.; Junpee, A.; Poovorawan, Y.; Scott, A.L. Detection and differentiation of filarial parasites by universal primers and polymerase chain reaction-restriction fragment length polymorphism analysis. Am. J. Trop. Med. Hyg. 2005, 73, 895–900. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; Bain, O.; Bourret, J.; Junker, K.; Guerrero, R.; Cañizales, I.; Kuzmin, Y.; Satoto, T.B.T.; Cardenas-Callirgos, J.M.; de Souza Lima, S.; et al. Shaking the tree: Multi-locus sequence typing usurps current Onchocercid (Filarial Nematode) phylogeny. PLoS Negl. Trop. Dis. 2015, 9, 1–19. [Google Scholar] [CrossRef]
- Gaillard, C.M.; Pion, S.D.; Hamou, H.; Sirima, C.; Bizet, C.; Lemarcis, T.; Rodrigues, J.; Esteban, A.; Peeters, M.; Mpoudi Ngole, E.; et al. Detection of DNA of filariae closely related to Mansonella perstans in faecal samples from wild non-human primates from Cameroon and Gabon. Parasit. Vectors 2020, 13, 313. [Google Scholar] [CrossRef]
- Ferri, E.; Barbuto, M.; Bain, O.; Galimberti, A.; Uni, S.; Guerrero, R.; Ferté, H.; Bandi, C.; Martin, C.; Casiraghi, M. Integrated taxonomy: Traditional approach and DNA barcoding for the identification of filarioid worms and related parasites (Nematoda). Front. Zool. 2009, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Casiraghi, M.; Anderson, T.J.C.; Bandi, C.; Bazzocchi, C.; Genchi, C. A phylogenetic analysis of filarial nematodes: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 2001, 1, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Taylor, M.J.; Hoerauf, A. Wolbachia bacteria of filarial nematodes. Parasitol. Today 1999, 15, 437–442. [Google Scholar] [CrossRef]
- Laidoudi, Y.; Ringot, D.; Watier-Grillot, S.; Davoust, B.; Mediannikov, O. A cardiac and subcutaneous canine dirofilariosis outbreak in a kennel in central France. Parasites 2019, 26, 72. [Google Scholar] [CrossRef]
- Rossi, M.I.D.; Aguiar-Alves, F.; Santos, S.; Paiva, J.; Bendas, A.; Fernandes, O.; Labarthe, N. Detection of Wolbachia DNA in blood from dogs infected with Dirofilaria immitis. Exp. Parasitol. 2010, 126, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Satjawongvanit, H.; Phumee, A.; Tiawsirisup, S.; Sungpradit, S.; Brownell, N.; Siriyasatien, P.; Preativatanyou, K. Molecular analysis of canine filaria and its Wolbachia endosymbionts in domestic dogs collected from two animal university hospitals in Bangkok metropolitan region, Thailand. Pathogens 2019, 8, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monica, A.; Matei, I.A.; Amico, G.D.; Bel, L.V.; Dumitrache, M.O.; Modrý, D.; Mihalca, A.D. Dirofilaria immitis and D. repens show circadian co-periodicity in naturally co-infected dogs. Parasit Vectors 2017, 10, 116. [Google Scholar]
- Mircean, M.; Ionică, A.M.; Mircean, V.; Györke, A.; Codea, A.R.; Tăbăran, F.A.; Taulescu, M.; Dumitrache, M.O. Clinical and pathological effects of Dirofilaria repens and Dirofilaria immitis in a dog with a natural co-infection. Parasitol. Int. 2017, 66, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, M.L.; Orihel, T.C. The genus Mansonella (syn. Tetrapetalonema): A new classification. Ann. Parasitol. Hum. Comp. 1984, 59, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Carme, B.; de Thoisy, B.; Motard, A.; Aznar, C.; Vié, J.C. Parasitoses humaines et mammifères sauvages en Guyane Française. Med. Trop. 2000, 60, 223–231. [Google Scholar]
- Vianna, L.M.M.; Martins, M.; Cohen, M.J.; Cohen, J.M.; Belfort, R. Mansonella ozzardi corneal lesions in the Amazon: A cross-sectional study. BMJ Open 2012, 2, e001266. [Google Scholar] [CrossRef] [Green Version]
- Garrido, C.; Campos, M. First report of presumed parasitic keratitis in Indians from the Brazilian Amazon. Cornea 2000, 19, 817–819. [Google Scholar] [CrossRef]
- Cohen, J.M.; Santana Ribeiro, J.A.; Martins, M. Ocular manifestations in mansonelliasis. Arq. Bras. Oftalmol. 2008, 71, 167–171. [Google Scholar]
- Baird, J.K.; Neafie, R.C.; Connor, D.H. Nodules in the conjunctiva, bung-eye, and bulge-eye in Africa caused by Mansonella perstans. Am. J. Trop. Med. Hyg. 1988, 38, 553–557. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.E.; Gárate, T.; Stavropoulos, C.; Fan, W.; González, L.M.; Eberhard, M.; Kimmelstiel, F.; Sordillo, E.M. Zoonotic filariasis caused by novel Brugia sp. Nematode, United States, 2011. Emerg. Infect. Dis. 2014, 20, 1248–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orihel, T.C.; Beaver, P.C. Zoonotic Brugia infections in North and South America. Am. J. Trop. Med. Hyg. 1989, 40, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Orihel, T.C.; Eberhard, M.L. Zoonotic filariasis. Clin. Microbiol. Rev. 1998, 11, 366–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalifoux, L.V. Filariasis, New World Primates. In Nonhuman Primates I. Monographs on Pathology of Laboratory Animals; Jones, T.C., Mohr, U., Hunt, R.D., Eds.; Springer: Berlin/Heidelberg, Germany, 1993; pp. 206–214. [Google Scholar]
- Notarnicola, J.; Jiménez, F.A.; Gardner, S.L. A new species of Dipetalonema (Filarioidea: Onchocercidae) from ateles chamek from the beni of Bolivia. J. Parasitol. 2007, 93, 661–667. [Google Scholar] [CrossRef] [Green Version]
- Notarnicola, J.; Pinto, C.M.; Navone, G.T. Host occurrence and geographical distribution of Dipetalonema spp. (Nematoda: Onchocercidae) in neotropical monkeys and the first record of Dipetalonema gracile in Ecuador. Comp. Parasitol. 2008, 75, 61–68. [Google Scholar] [CrossRef]
- Chabaud, A.G. Le genre Dipetalonema Diesing 1861; Essai de classification. Ann. Parasitol. Hum. Comp. 1952, 27, 250–285. [Google Scholar] [CrossRef] [Green Version]
- Bain, O.; Petit, G.; Rosales-Loesener, L. Filaires de singes sud-américains. Bull. Muséum Natl. Histoire Nat. Sect. A Zool. Biol. Ecologie Anim. 1986, 8, 513–542. [Google Scholar]
- Bain, O.; Diagne, M.; Muller, R. Une cinquième filaire du genre Dipetalonema, parasite de singes sud-américains. Ann. Parasitol. Hum. Comp. 1987, 62, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Esslinger, J.H. Dipetalonema obtusa (McCoy, 1936) comb. n. (Filarioidea: Onchocercidae) in colombian primates, with a description of the adult. J. Parasitol. 1966, 52, 498–502. [Google Scholar] [CrossRef]
- Vanderhoeven, E.A.; Notarnicola, J.; Agostini, I. First record of Dipetalonema robini Petit, Bain & Roussilhon 1985 (Nematoda: Onchocercidae) parasitizing sapajus nigritus in northeastern argentina. Mastozoologia Neotropical. 2017, 24, 483–488. [Google Scholar]
- UICN France. La Liste rouge des espèces menacées en France; Muséum National d’Histoire Naturelle: Paris, France, 2015; p. 12. [Google Scholar]
- Boubli, J.P.; di Fiore, A.; Mittermeier, R.A.; dos Santos, M. Alouatta macconnelli. The IUCN Red List of Threatened Species 2020, e.T40642A17926817. Downloaded on 22 July 2020. [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial Cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Meyer, C.P. Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biol. J. Linn. Soc. 2003, 79, 401–459. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Fong, M.Y.; Noordin, R.; Lau, Y.L.; Cheong, F.W.; Yunus, M.H.; Idris, Z.M. Comparative analysis of ITS1 nucleotide sequence reveals distinct genetic difference between Brugia malayi from Northeast Borneo and Thailand. Parasitology 2013, 140, 39–45. [Google Scholar] [CrossRef] [Green Version]
- McNulty, S.N.; Mitreva, M.; Weil, G.J.; Fischer, P.U. Inter and intra-specific diversity of parasites that cause lymphatic filariasis. Infect. Genet. Evol. 2013, 14, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 2012, 13, 134. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Windsor, D.M. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. R. Soc. B Biol. Sci. 2000, 267, 1277–1285. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [Green Version]







| Sample Code | Filarial DNA | Wolbachia DNA | Decision | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Filariids | ITS genus-specific PCRs | Wolbachia 16S-specific PCRs | |||||||
| 28S qPCR | 18S PCR | COI PCR | Mansonella spp. PCR | Brugia spp. PCR | Brugia spp. qPCR | Wolbachia 16S qPCR | Wolbachia 16S PCR | Combined Assays | |
| B1 | N/A | N/A | N/A | N/A | N/A | Neg. | Neg. | N/A | Negative. |
| B2 | Pos. | Mansonella sp. [MT336169] | O/P | Mansonella sp. [MT341515] | N/A | Pos. | Neg. | N/A | Mansonella sp. + Brugia sp. |
| B3 | Pos. | Mansonella sp. [MT336170] | O/P | Mansonella sp. [MT341516] | Brugia sp. [MT341511] | Pos. | Pos. | O/P | Mansonella sp. + Brugia sp. |
| B4 | Pos. | Mansonella sp. [MT336171] | O/P | Mansonella sp. [MT341517] | Brugia sp. [MT341512] | Pos. | Pos. | W-Mansonella sp. [MT231961] | Mansonella sp. + Brugia sp. |
| B5 | Pos. | unidentified Onchocercidae species [MT336175] | O/P | Mansonella sp. [MT341518] | N/A | Neg. | Pos. | W-Mansonella sp. [MT231962] | Mansonella sp. + unidentified Onchocercidae species |
| B6 | Pos. | Mansonella sp. [MT336172] | O/P | Mansonella sp. [MT341519] | N/A | Neg. | Pos. | W-Brugia sp. [MT231964] | Mansonella sp. + Brugia sp. |
| B7 | Pos. | Mansonella sp. [MT336173] | O/P | Mansonella sp. [MT341520] | Brugia sp. [MT341513] | Pos. | Neg. | N/A | Mansonella sp. + Brugia sp. |
| B8 | Pos. | Mansonella sp. [MT336174] | Mansonella sp. [MT724663] | Mansonella sp. [MT341521] | N/A | Neg. | Pos. | W-Mansonella sp. [MT231963] | Mansonella sp. |
| B9 | Pos. | Brugia sp. [MT336168] | Brugia sp. [MT724693] | N/A | Brugia sp. [MT341514] | Pos. | Pos. | W-Brugia sp. [MT231965] | Brugia sp. |
| Genera | Species | Host | References |
|---|---|---|---|
| Mansonella (Faust, 1929), Mansonella (Tetrapetalonema) comb. n. (Faust 1935) | Mansonella (T.) marmosetae (Faust 1935) | Saguinus geoffroyi, Saimiri oerstedii oerstedii, Ateles paniscus, Saimiri boliviensis, Saimiri sciureus and Alouatta spp. | [44,66,67,76] |
| Mansonella (T.) zakii (Nagaty 1935) | Leontopithecus (= Leontocebus) rosalia | ||
| Mansonella (T.) panamensis (McCoy 1936) | Cebus capucinus, Saimiri oerstedii oerstedii, Aotus lemurinus zonalis, C. apella and A. trivirgatus | ||
| Mansonella (T.) atelensis atelensis (McCoy 1935) | Ateles geoffroyi, A. fusciceps rufiventris | ||
| Mansonella (T.) atelensis amazonae (Bain and Guerrero 2015) | Cebus olivaceus | ||
| Mansonella (T.) parvum (McCoy 1936) | Cebus capucinus, Saimiri oerstedii oerstedii | ||
| Mansonella (T.) obtusa (McCoy 1936) | Cebus capucinus, C. capucinus, C. albifrons, Saimiri oerstedii oerstedii | ||
| Mansonella (T.) tamarinae (Dunn and Lambrecht 1963) | Saguinus (= Tamarinus) nigricollis | ||
| Mansonella (T.) barbascalensis (Esslinger and Gardiner 1974) | Aotus trivirgatus | ||
| Mansonella (T.) mystaxi (Eberhard 1978) | Saguinus mystax mystax | ||
| Mansonella (T.) saimiri (Esslinger 1981) | Saimiri sciureus | ||
| Mansonella (T.) peruviana (Bain, Petit and Rosales-Loesener 1986) | Saimiri sciureus | ||
| Mansonella (T.) colombiensis (Esslinger 1982) | Saimiri sciureus, Cebus apella | ||
| Mansonella (T.) mariae (Petit, Bain and Roussilhon 1985) | Saimiri sciureus | ||
| Dipetalonema (Diesing 1861) | D. gracile (Rudolphi 1819) | Saimiri sciureus, Cebus albifrons, A. geoffroyi, Aotus lemurinus, Ateles chamek, Ateles fusciceps, Ateles geoffroyi, Ateles paniscus, Cebus apella, Cebus capucinus, Cebus spp., Lagothrix lagothricha, Saguinus mystax, Saguinus nigricollis, Saimiri oerstedii, Saimiri sciureus, Saimiri sciureus, Sapajus macrocephalus, B. arachnoïdes, L. rosalia, Leontopithecus chrysopygus, Saguinus bicolor, Cebus albifrons | [76,77,78,79,80,81,82] |
| D. graciliformis (Freitas 1964) | Saguinus midas | ||
| D. robini (Petit et al. 1985) | Saimiri sciureus, Sapajus nigritus, Saimiri boliviensis, Cebus spp. | ||
| D. freitasi (Bain, Diagne and Muller 1987) | Cebus capucinus | ||
| D. caudispina (Molin 1858) | Alouatta seniculus, Ateles paniscus, Brachyteles arachnoides, Cebus albifrons, Cebus apella, Lagothrix lagotricha, Leontopithecus rosalia, Saimiri sciureus, Saimiri sciureus, Sapajus macrocephalus | ||
| D. obtusa (McCoy 1936) | Cebus albifron, Cebus capucinus | ||
| D. yatesi (Julians 2007) | Ateles chamek |
| System Name | Target Gene | Primer and Probe Name | Sequence (5′–3′) | Amplicon Size (bp) | Tm/Elongation Time | Assay Specificity | Ref. |
|---|---|---|---|---|---|---|---|
| Pan-fil 28S qPCR-based system | LSU rRNA (28S) | qFil-28S-F | TTGTTTGAGATTGCAGCCCA | 151 | 60 °C/30” | Filariids | [24] |
| qFil-28S-P | 6FAM-CAAGTACCGTGAGGGAAAGT-TAMRA | ||||||
| qFil-28S-R | GTTTCCATCTCAGCGGTTTC | ||||||
| All-Wol 16S qPCR-based system | 16S rRNA gene | all.Wol.16S.301-F | TGGAACTGAGATACGGTCCAG | 177 | 61 °C/30” | Wolbachia | |
| all.Wol.16S.347-P | 6FAM-AATATTGGACAATGGGCGAA-TAMRA | ||||||
| all.Wol.16S.478-R | GCACGGAGTTAGCCAGGACT | ||||||
| 16S W-Spec | W-Specf | CATACC TATTCGAAGGGATAG | 438 | 60 °C/1’ | [89] | ||
| W-Specr | AGCTTCGAGTGAA ACCAATTC | ||||||
| Brug-gen-spec qPCR | Internal Transcribed Spacer 1 (ITS1) | Brug.ITS.f.260 | AGCGATAGCTTAATTAATTTTACCATTT | 161 | 61 °C/30” | Brugia spp. | This study |
| Brug.ITS.p.307 | 6FAM- GCATTTATGCTAGATATGCTACCAA-TAMRA | ||||||
| Brug.ITS.r.421 | CCACCGCTAAGAGTTAAAAAAATT | ||||||
| Brug-gen-spec PCR | Fil.ITS.f: | GAACCTGCGGAAGGATCA | 417–441 | 54 °C/30” | |||
| Brug.ITS.r | CCACCGCTAAGAGTTAAAAAAATT | ||||||
| Manso-gen-spec PCR | Fil.ITS.f: | GAACCTGCGGAAGGATCA | 333–345 | 55 °C/30” | Mansonella spp. | ||
| Manso.ITS.r | TGTGTATTTATTTGTTGGTAGCATATT | ||||||
| SSU rRNA (18S) | Fwd.18S.631 | TCGTCATTGCTGCGGTTAAA | 1127–1155 | 54 °C/1’30” | Nematoda | [61] | |
| Rwd.18S.1825r | GGTTCAAGCCACTGCGATTAA | ||||||
| Pan-fil cox1 PCR | Cytochrome c oxidase subunit 1 gene (cox1) | Fwd.957 | ATRGTTTATCAGTCTTTTTTTATTGG | 509 | 52 °C/1’ | Filariids | [24] |
| Rwd.1465 | GCAATYCAAATAGAAGCAAAAGT | ||||||
| dg-Folmer’s primers | dgLCO-1490 | GGTCAACAAATCATAAAGAYATYGG | 708 | 44 °C/40” | Metazoans | [86] | |
| dgHCO-2198 | TAAACTTCAGGGTGACCAAARAAYCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laidoudi, Y.; Medkour, H.; Levasseur, A.; Davoust, B.; Mediannikov, O. New Molecular Data on Filaria and its Wolbachia from Red Howler Monkeys (Alouatta macconnelli) in French Guiana—A Preliminary Study. Pathogens 2020, 9, 626. https://doi.org/10.3390/pathogens9080626
Laidoudi Y, Medkour H, Levasseur A, Davoust B, Mediannikov O. New Molecular Data on Filaria and its Wolbachia from Red Howler Monkeys (Alouatta macconnelli) in French Guiana—A Preliminary Study. Pathogens. 2020; 9(8):626. https://doi.org/10.3390/pathogens9080626
Chicago/Turabian StyleLaidoudi, Younes, Hacène Medkour, Anthony Levasseur, Bernard Davoust, and Oleg Mediannikov. 2020. "New Molecular Data on Filaria and its Wolbachia from Red Howler Monkeys (Alouatta macconnelli) in French Guiana—A Preliminary Study" Pathogens 9, no. 8: 626. https://doi.org/10.3390/pathogens9080626
APA StyleLaidoudi, Y., Medkour, H., Levasseur, A., Davoust, B., & Mediannikov, O. (2020). New Molecular Data on Filaria and its Wolbachia from Red Howler Monkeys (Alouatta macconnelli) in French Guiana—A Preliminary Study. Pathogens, 9(8), 626. https://doi.org/10.3390/pathogens9080626