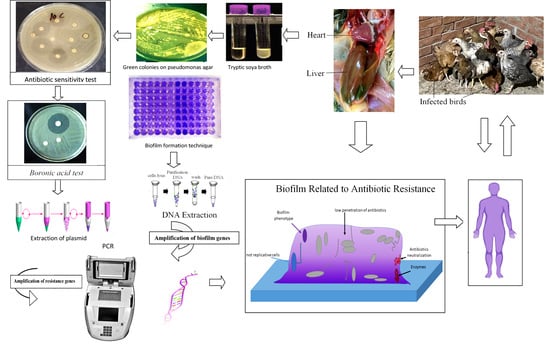
Detection of β-Lactamase Resistance and Biofilm Genes in Pseudomonas Species Isolated from Chickens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Consent for Participation
2.2. Ethical Approval
2.3. ARRIVE Principles
2.4. Bacterial Strains
2.5. Antimicrobial Susceptibility Testing
2.6. Phenotypic and Genotypic Screening for AmpC β-Lactamases
2.6.1. Screening for AmpC β-lactamases
2.6.2. Confirmation of AmpC β-lactamase Production
Boronic Acid Test
Disk Test
2.6.3. PCR Technique for the Detection of Antimicrobial Resistance Genes
Extraction of Plasmid-Mediated AmpC β-lactamases
Multiplex PCR Testing for the blaCMY and blaMIR
Uniplex PCR Testing for DHA and FOX
2.6.4. Biofilm Detection Techniques
Microtiter Plate Technique
P. aeruginosa ATCC 27853 Positive Reference Strain for Biofilm Formation
2.6.5. PCR Technique for Biofilm Formation Gene Detection
DNA Extraction
Uniplex PCR Reaction
2.6.6. Essential Oil Chromatography
Clove Oil
Gas Chromatography–Mass Spectrometry (GC-MS) Analysis of Clove Oil
2.6.7. Screening Antibiofilm Effects of Clove Oil on Pseudomonas spp.
Antibiofilm Activity of Clove Oil
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kebede, F. Pseudomonas infection in chickens. J. Vet. Med. Anim. Health 2010, 2, 55–58. [Google Scholar]
- Tartor, Y.H.; El-Naenaeey, E.Y. RT-PCR detection of exotoxin genes expression in multidrug resistant Pseudomonas aeruginosa. Cell. Mol. Biol. 2016, 62, 56–62. [Google Scholar]
- Hinton, A.; Cason, J.A.; Ingram, K.D. Tracking spoilage bacteria in commercial poultry processing and refrigerated storage of poultry carcasses. Int. J. Food Microbiol. 2004, 91, 155–165. [Google Scholar] [CrossRef]
- Morales, P.A.; Aguirre, J.S.; Troncoso, M.R.; Figueroa, G.O. Phenotypic and genotypic characterization of Pseudomonas spp. present in spoiled poultry fillets sold in retail settings. LWT Food Sci. Technol. 2016, 73, 609–614. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, A.; Mena, A.; Maciá, M.D. Evolution of Pseudomonas aeruginosa Pathogenicity: From Acute to Chronic Infections. In Evolutionary Biology of Bacterial and Fungal Pathogens; Baquero, F., Nombela, C., Cassell, G.H., Gutiérrez-Fuentes, J.A., Eds.; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; Walker, N.; Scales, B.S.; Beck, J.M.; Huffnagle, G.B. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS ONE 2014, 9, e97214. [Google Scholar] [CrossRef] [Green Version]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef] [Green Version]
- Slama, T.G. Gram-negative antibiotic resistance: There is a price to pay. Crit. Care 2008, 12 (Suppl. 4), S4. [Google Scholar] [CrossRef] [Green Version]
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A.; Petersen, P.J.; Fingerman, I.M.; White, D.G. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J. Antimicrob. Chemother. 1999, 44, 607–610. [Google Scholar] [CrossRef]
- Gniadkowski, M. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin. Microbiol. Infect. 2001, 7, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Karballaei Mirzahosseini, H.; Hadadi-Fishani, M.; Morshedi, K.; Khaledi, A. Meta-analysis of biofilm formation, antibiotic resistance pattern, and biofilm-related genes in Pseudomonas aeruginosa isolated from clinical samples. Microb. Drug Resist. 2020, 26, 815–824. [Google Scholar] [CrossRef]
- Ejikeugwu, C.; Nworie, O.; Saki, M.; Al-Dahmoshi, H.O.M.; Al-Khafaji, N.S.K.; Ezeador, C.; Nwakaeze, E.; Eze, P.; Oni, E.; Obi, C.; et al. Metallo-b-lactamase and AmpC genes in Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolates from abattoir and poultry origin in Nigeria. BMC Microbiol. 2021, 21, 124. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, L.Z. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013, 14, 20983–21005. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 2011, 34, 737–751. [Google Scholar] [CrossRef]
- Aryal, M.; Muriana, P.M. Efficacy of commercial sanitizers used in food processing facilities for inactivation of Listeria monocytogenes, E. coli O157: H7, and Salmonella biofilms. Foods 2019, 8, 639. [Google Scholar] [CrossRef] [Green Version]
- Mith, H.; Dure, R.; Delcenserie, V.; Zhiri, A.; Daube, G.; Clinquart, A. Antimicrobial activities of commercial essential oils and their components against food-borne pathogens and food spoilage bacteria. Food Sci. Nutr. 2014, 2, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Zhai, H.; Liu, H.; Wang, S.; Wu, J.; Kluenter, A.M. Potential of essential oils for poultry and pigs. Anim. Nutr. 2018, 4, 179–186. [Google Scholar] [CrossRef]
- Chaieb, K.; Hajlaoui, H.; Zmantar, T.; Kahla-Nakbi, A.B.; Rouabhia, M.; Mahdouani, K.; Bakhrouf, A. The chemical composition and biological activity of clove essential oil, Eugenia caryophyllata (Eugenia caryophyllata L. Myrtaceae): A short review. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2007, 21, 501–506. [Google Scholar]
- Heredia-Guerrero, J.A.; Ceseracciu, L.; Guzman-Puyol, S.; Paul, U.C.; Alfaro-Pulido, A.; Grande, C.; Bayer, I.S. Antimicrobial, antioxidant, and waterproof RTV silicone-ethyl cellulose composites containing clove essential oil. Carbohydr. Polym. 2018, 192, 150–158. [Google Scholar] [CrossRef]
- Helander, I.M.; Alakomi, H.L.; Latva-Kala, K.; Mattila-Sandholm, T.; Pol, I.; Smid, E.J.; von Wright, A. Characterization of the action of selected essential oil components on Gram-negative bacteria. J. Agric. Food Chem. 1998, 46, 3590–3595. [Google Scholar] [CrossRef]
- Zhou, L.; Zheng, H.; Tang, Y.; Yu, W.; Gong, Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol. Lett. 2013, 35, 631–637. [Google Scholar] [CrossRef]
- Vijayasteltar, L.; Nair, G.G.; Maliakel, B.; Kuttan, R.; Krishnakumar, I.M. Safety assessment of a standardized polyphenolic extract of clove buds: Subchronic toxicity and mutagenicity studies. Toxicol. Rep. 2016, 3, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Du Sert, N.P.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Würbel, H. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 2020, 18, e3000411. [Google Scholar]
- Shahat, H.S.; Mohamed, H.; Al-Azeem, A.; Mohammed, W.; Nasef, S.A. Molecular detection of some virulence genes in Pseudomonas aeruginosa isolated from chicken embryos and broilers with regard to disinfectant resistance. SVU-Int. J. Vet. Sci. 2019, 2, 52–70. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Bogaerts, P.; Deplano, A.; Berhin, C.; Huang, T.D.; Van Eldere, J.; Rodriguez-Villalobos, H. Detection and characterization of class A extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J. Antimicrob. Chemother. 2010, 65, 866–871. [Google Scholar] [CrossRef] [Green Version]
- Legakis, N.J.; Aliferopoulou, M.; Papavassiliou, J.; Papapetropoulou, M. Serotypes of Pseudomonas aeruginosa in clinical specimens in relation to antibiotic susceptibility. J. Clin. Microbiol. 1982, 16, 458–463. [Google Scholar] [CrossRef] [Green Version]
- John, D.T.; James, H.J.; Murray, P.R.; Baron, E.J.; Pfaller, M.A.; Tenover, F.C.; Morton, D. Manual of Clinical Microbiology; ASM Publishers: Washington, DC, USA, 2009; pp. 302–305. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing [EUCAST]. Breakpoints Tables for Interpretation of MICs and Zone Diameter; Version 8.0.; EUCAST: Växjö, Sweden, 2018. [Google Scholar]
- Tan, T.Y.; Ng, S.Y.; Teo, L.; Koh, Y.; Teok, C.H. Detection of plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J. Clin. Pathol. 2008, 61, 642–644. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Matten, J.; Rabsch, W. Salmonella enterica serovar Typhi with CTX-M β-lactamase, Germany. Emerg. Infect. Dis. 2009, 15, 1533. [Google Scholar] [CrossRef]
- Reuland, E.A.; Halab, T.; Hays, J.P.; de Jongh, D.M.; Snetselaar, H.D.; van Keulen, M.; Elders, P.J.; Savelkoul, P.H.; Vandenbroucke-Grauls, C.M.; Al Naiemi, N. Plasmid-mediated AmpC: Prevalence in community-acquired isolates in Amsterdam, the Netherlands, and risk factors for carriage. PLoS ONE 2015, 10, e0113033. [Google Scholar] [CrossRef]
- Hou, W.; Sun, X.; Wang, Z.; Zhang, Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5624–5631. [Google Scholar] [CrossRef] [Green Version]
- Wassef, M.; Behiry, I.; Younan, M.; El Guindy, N.; Mostafa, S.; Abada, E. Genotypic Identification of AmpC β-Lactamases Production in Gram-Negative Bacilli Isolates. Jundishapur J. Microbiol. 2014, 7, e8556. [Google Scholar] [CrossRef] [Green Version]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179.43. [Google Scholar] [CrossRef]
- Tawakol, M.; Nehal, N.; Reem, R. Molecular studies on some virulence factors of Pseudomonas aeruginosa isolated from chickens as a biofilm forming bacteria. Assiut Vet. Med. J. 2018, 64, 43–51. [Google Scholar]
- Kerekes, E.B.; Vidács, A.; Takó, M.; Petkovits, T.; Vágvölgyi, C.; Horváth, G.; Krisch, J. Anti-biofilm effect of selected essential oils and main components on mono-and polymicrobic bacterial cultures. Microorganisms 2019, 7, 345. [Google Scholar] [CrossRef] [Green Version]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Dutta, T.K.; Roychoudhury, P.; Samanta, I.; Kalai, S.; Bandyopadhyay, S. Molecular characterization of biofilm-producing Pseudomonas aeruginosa isolated from healthy pigs and Chicken in India. Indian J. Anim. Res. 2020, 54, 1400–1407. [Google Scholar] [CrossRef] [Green Version]
- Blair, J.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.D.; Okuthe, G.E.; Apalata, T. Detection of extended spectrum beta-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci. Rep. 2021, 11, 7110. [Google Scholar] [CrossRef]
- Mohanty, S.; Baliyarsingh, B.; Nayak, S.K. Antimicrobial Resistance in Pseudomonas aeruginosa: A concise review. In Antimicrobial Resistance—A One Health Perspective; IntechOpen: London, UK, 2020. [Google Scholar]
- Thomson, K.S. Extended-spectrum-beta-lactamase, AmpC, and Carbapenemase issues. J. Clin. Microbiol. 2010, 48, 1019–1025. [Google Scholar] [CrossRef] [Green Version]
- Black, J.A.; Moland, E.S.; Thomson, K.S. AmpC disk test for detection of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal AmpC β-lactamases. J. Clin. Microbiol. 2005, 43, 3110–3113. [Google Scholar] [CrossRef]
- Polsfuss, S.; Bloemberg, G.V.; Giger, J.; Meyer, V.; Böttger, E.C.; Hombach, M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–80351. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [Green Version]
- Helmy, M.M.; Wasfi, R. Phenotypic and molecular characterization of plasmid mediated AmpC β-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. Biomed Res. Int. 2014, 2014, 171548. [Google Scholar] [CrossRef] [Green Version]
- Wendorf, K.A.; Kay, M.; Baliga, C.; Weissman, S.J.; Gluck, M.; Verma, P.; D’Angeli, M.; Swoveland, J.; Kang, M.G.; Eckmann, K.; et al. Endoscopic retrograde cholangiopancreatography-associated AmpC Escherichia coli outbreak. Infect. Control Hosp. Epidemiol. 2015, 36, 634–642. [Google Scholar] [CrossRef] [Green Version]
- Haldorsen, B.; Aasnaes, B.; Dahl, K.H.; Hanssen, A.M.; Simonsen, G.S.; Walsh, T.R.; Sundsfjord, A.; Lundblad, E.W. The AmpC phenotype in Norwegian clinical isolates of Escherichia coli is associated with an acquired ISEcp1-like AmpC element or hyperproduction of the endogenous AmpC. J. Antimicrob. Chemother. 2008, 62, 694–702. [Google Scholar] [CrossRef] [Green Version]
- Fam, N.S.; Gamal, D.; Said, M.E.; Aboul-Fadl, L.; Dabei, E.E.; Attar, S.E.; Sorur, A.; Fouad, S.A.; Klena, J.D. Detection of plasmid-mediated AmpC beta-lactamases in clinically significant bacterial isolates in a research institute hospital in Egypt. Life Sci. J. 2013, 10, 2294–2304. [Google Scholar]
- Hosny, A.E.-D.M.S.; Mona, T.K. A study on occurrence of plasmid mediated AmpC ß-lactamases among gram negative clinical isolates and evaluation of different methods used for their detection. J. Appl. Sci. Res. 2012, 8, 2280–2285. [Google Scholar]
- Wollheim, C.; Guerra, I.M.; Conte, V.D.; Hoffman, S.P.; Schreiner, F.J.; Delamare, A.P.; Barth, A.L.; Echeverrigaray, S.; Costa, S.O. Nosocomial and community infections due to class A extended-spectrum β-lactamase (ESBLA)-producing Escherichia coli and Klebsiella spp. in southern Brazil. Braz. J. Infect. Dis. 2011, 15, 138–143. [Google Scholar]
- Sommer, L.M.; Johansen, H.K.; Molin, S. Antibiotic resistance in Pseudomonas aeruginosa and adaptation to complex dynamic environments. Microb. Genom. 2020, 6, e000370. [Google Scholar] [CrossRef]
- Fadare, F.T.; Okoh, A.I. Distribution and molecular characterization of ESBL, pAmpC β-lactamases, and non-β-lactam encoding genes in Enterobacteriaceae isolated from hospital wastewater in Eastern Cape Province, South Africa. PLoS ONE 2021, 16, e0254753. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect Control 2019, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Klausen, M.; Heydorn, A.; Ragas, P.; Lambertsen, L.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003, 48, 1511–1524. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Salama, Y.; Chennaoui, M.; Sylla, A.; Mountadar, M.; Rihani, M.; Assobhei, O. Characterization, structure, and function of extracellular polymeric substances (EPS) of microbial biofilm in biological wastewater treatment systems: A review. Desalination Water Treat. 2016, 57, 16220–16237. [Google Scholar] [CrossRef]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.S.A.; Ahmad, I.; Sajid, M.; Cameotra, S.S. Current and Emergent Control Strategies for Medical Biofilms. In Antibiofilm Agents; Rumbaugh, K., Ahmad, I., Eds.; Springer Series on, Biofilms; Springer: Berlin/Heidelberg, Germany, 2014; Volume 8. [Google Scholar]
- Empel, J.; Baraniak, A.; Literacka, E.; Mrówka, A.; Fiett, J.; Sadowy, E.; Hryniewicz, W.; Gniadkowsk, M. Beta-PL Study Group. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 2008, 52, 2449–2454. [Google Scholar] [CrossRef] [Green Version]
- Banar, M.; Emaneini, M.; Satarzadeh, M.; Abdellahi, N.; Beigverdi, R.; Leeuwen, W.B.; Jabalameli, F. Evaluation of Mannosidase and Trypsin Enzymes Effects on Biofilm Production of Pseudomonas aeruginosa Isolated from Burn Wound Infections. PLoS ONE 2016, 11, e0164622. [Google Scholar] [CrossRef] [Green Version]
- Karami, P.; Khaledi, A.; Mashoof, R.Y.; Yaghoobi, M.H.; Karami, M.; Dastan, D.; Alikhani, M.Y. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Rep. 2020, 18, 100561. [Google Scholar] [CrossRef]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 2006, 188, 8213–8221. [Google Scholar] [CrossRef] [Green Version]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [Green Version]
- Ghadaksaz, A.; Fooladi, A.A.I.; Hosseini, H.M.; Amin, M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J. Appl. Biomed. 2015, 13, 61–68. [Google Scholar] [CrossRef]
- Olson, M.E.; Ceri, H.; Morck, D.W.; Buret, A.G.; Read, R.R. Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002, 66, 86. [Google Scholar]
- Sala, C.; Morar, A.; Colibar, O.; Morvay, A.A. Antibiotic resistance of gram negative bacteria isolated from meat surface biofilm. Rom Biotech. Lett. 2012, 17, 7483–7492. [Google Scholar]
- Abidi, S.H.; Sherwani, S.K.; Siddiqui, T.R.; Bashir, A.; Kazmi, S.U. Drug resistance profile and biofilm forming potential of Pseudomonas aeruginosa isolated from contact lenses in Karachi-Pakistan. BMC Ophthalmol. 2013, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Corehtash, Z.G.; Khorshidi, A.; Firoozeh, F.; Akbari, H.; Aznaveh, A.M. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J. Microbiol. 2015, 8, e22345. [Google Scholar]
- Behzadi, P.; Ambrosi, C.; Scribano, D.; Zanetti, S.; Sarshar, M.; Gajdács, M.; Donadu, M.G. Editorial: Current perspectives on Pseudomonas aeruginosa: Epidemiology, virulence and contemporary strategies to combat multidrug-resistant (MDR) pathogens. Front Microbiol. 2022, 13, 975616. [Google Scholar] [CrossRef]
- Cao, H.; Xia, T.; Li, Y.; Xu, Z.; Bougouffa, S.; Lo, Y.K.; Bajic, V.B.; Luo, H.; Woo, P.C.Y.; Yan, A. Uncoupled Quorum Sensing Modulates the Interplay of Virulence and Resistance in a Multidrug-Resistant Clinical Pseudomonas aeruginosa Isolate Belonging to the MLST550 Clonal Complex. Antimicrob. Agents Chemother. 2019, 63, e01944-18. [Google Scholar] [CrossRef] [Green Version]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef] [PubMed]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar] [CrossRef]
- Kavanaugh, N.L.; Ribbeck, K. Selected antimicrobial essential oils eradicate Pseudomonas spp. and Staphylococcus aureus biofilms. Appl. Environ. Microbiol. 2012, 78, 4057–4061. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, Y.; Zhu, X.; Cao, P.; Wei, S.; Lu, Y. Antibacterial and antibiofilm activities of eugenol from essential oil of Syzygium aromaticum (L.) Merr. & LM Perry (clove) leaf against periodontal pathogen Porphyromonas gingivalis. Microb. Pathog. 2017, 113, 396–402. [Google Scholar] [PubMed]
- Kim, Y.G.; Lee, J.H.; Gwon, G.; Kim, S.I.; Park, J.G.; Lee, J. Essential oils and eugenols inhibit biofilm formation and the virulence of Escherichia coli O157: H7. Sci. Rep. 2016, 6, 36377. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.; Gilbert, E.S. Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 2004, 70, 6951–6956. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Du, M.; Fan, M.; Bian, Z. In vitro activity of eugenol against Candida albicans biofilms. Mycopathologia 2007, 163, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Yadav, E. A phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PLoS ONE 2015, 10, e0119564. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, H.M.; Emeish, W.F.; Braeuning, A.; Hammad, S. Detection of aflatoxin-producing fungi isolated from Nile tilapia and fish feed. EXCLI J. 2017, 16, 1308. [Google Scholar]
- Perez-Conesa, D.; McLandsborough, L.; Weiss, J. Inhibition and inactivation of Listeria monocytogenes and Escherichia coli O157: H7 colony biofilms by micellar-encapsulated eugenol and carvacrol. J. Food Prot. 2006, 69, 2947–2954. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]






| Genes | Primer Sequence (5′–3′) | Amplicon Size (bp) | Annealing Temperature °C | References |
|---|---|---|---|---|
| bla CMY | F-5′-TGG CCA GAA CTG ACA GGC AAA-3′ | 462 | 60 | [38] |
| R-5′-TTT CTC CTG AAC GTG GCT GGC-3′ | ||||
| bla MIR | F-5′-TCG GTA AAG CCG ATG TTG CGG-3′ | 302 | 60 | |
| R-5′-TTT CTC CTG AAC GTG GCT GCT GGC-3′ | ||||
| DHA | F-5′-AAC TTT CAC AGG TGT GCT GGG T-3′ | 405 | 55 | |
| R-5′-CCG TAC GCA TAC TGG CTT TGC-3′ | ||||
| FOX | F-5′-AAC ATG GGG TAT CAG GGA GAT G-3′ | 190 | 55 | |
| R-5′-CAA AGC GCG TAA CCG GAT TGG-3′ | ||||
| pslD | F-5′-TGTACACCGTGCTCAACGAC-3′ | 369 | 58 | [39] |
| R-5′-CTTCCGGCCCGATCTTCATC-3′ | ||||
| rhlA | F:5′-TGCTGATGGTTGCTGGCTTTC-3′ | 89 | 58 | |
| R:5′-CTCGGTGGTGATGGCATTCG-3′ | ||||
| pelA | F:5′-CATACCTTCAGCCATCCGTTCTTC-3′ | 786 | 60 | [40] |
| R:5′-CGCATTCGCCGCACTCAG-3′ |
| Pseudomonas Isolates | No. (%) Strong Biofilm Producer Isolates | No. (%) Moderate Biofilm Producer Isolates | No. (%) Weak/Non Biofilm Producer Isolates |
|---|---|---|---|
| P. aeruginosa (n = 7) | 4 (57.14%) | 3 (42.86%) | - |
| P.fluorescens (n = 11) | 4 (36. 36%) | 4 (36.36%) | 3 (27.27%) |
| Total (18) | 8 (44.4%) | 7 (38.8%) | 3 (16.6%) |
| Antibiotic | Bacterial Strains | Biofilm Formation Positive | Biofilm Formation Negative | Total Resistance Strains (%) |
|---|---|---|---|---|
| Piperacillin | P. aeruginosa | 4 (57.1%) | ------- | 4 (57.1) |
| P. fluorescens | 4 (36.3%) | 3 (27.2%) | 7 (63.6) | |
| Levofloxacin | P. aeruginosa | ----- | --- | ------ |
| P. fluorescens | 3 (27.2%) | 1 (9.1%) | 4 (36.3) | |
| Ciprofloxacin | P. aeruginosa | ------ | ----- | ----- |
| P. fluorescens | 7 (63.6%) | 1 (9%) | 8 (72.7) | |
| Gentamicin | P. aeruginosa | 1 (14.2%) | ------ | 1(14.2) |
| P. fluorescens | 5 (45.4%) | 3 (27.2%) | 8 (72.7) | |
| Cefotaxime | P. aeruginosa | 7 (100%) | ------- | 7 (100) |
| P. fluorescens | 8 (72.7%) | 3 (27.2) | 11 (100) | |
| Cefazoline | P. aeruginosa | 7 (100%) | ----------- | 7 (100) |
| P. fluorescens | 8 (72.7%) | 3 (27.2) | 11 (100) | |
| Imipenem | P. aeruginosa | 4 (57.2%) | ------- | 4 (57.2) |
| P. fluorescens | 5 (45.5%) | 3 (27.27%) | 8 (72.7) | |
| Meropenem | P. aeruginosa | 5 (71.4%) | ------ | 5 (71.4) |
| P. fluorescens | 6 (54.5.4%) | 2 (18.1%) | 8 (72.7) | |
| Norfloxacin | P. aeruginosa | 0 (0%) | --- | 2 (28.5) |
| P. fluorescens | 4 (36.3%) | 2 (18.1%) | 6 (54.5) | |
| Amikacin | P. aeruginosa | 6 (85.7%) | --- | 6 (85.7) |
| P. fluorescens | 8 (72.7%) | 3 (27.27%) | 11 (100) | |
| Tetracyclines | P. aeruginosa | 7 (100%) | ----- | 7 (100) |
| P. fluorescens | 8 (72.7%) | 3 (27.2) | 11 (100) | |
| Chloramphenicol | P. aeruginosa | 3 (42.8%) | --- | 3 (42.8) |
| P. fluorescens | 8 (72.72%) | 3 (27.27%) | 11 (100) |
| Gene | P. aeruginosa (No = 7) | P. fluorescens (No = 11) | Total (No. of Isolates = 18) |
|---|---|---|---|
| Biofilm genes | |||
| PslD | 6 (85.7%) | 9 (81.8%) | 15 (83.3%) |
| pelA | 4 (57.1%) | 2 (18.1%) | 6 (40%) |
| rhlA | 5 (71.4%)) | 9 (81.8%) | 14 (77.8%) |
| AmpC β-Lactamases genes | |||
| blaCMY-type genes | 7 (100%) | 11 (100%) | 18 (100%) |
| blaMIR-type genes | ---- | 3 (27.2%) | 3 (16.6%) |
| DHA | 3 (42.8%) | 2 (18.1%) | 5 (27.7%) |
| FOX | - | - | - |
| Peak | R.t * | Name | Area % | Molecular Weight | Molecular Formula | MF ** |
|---|---|---|---|---|---|---|
| 1 | 8.34 | Benzyl alcohol | 37.12 | 108 | C7H8O | 958 |
| 2 | 8.98 | PHENOL, 2-METHYL | 1.07 | 108 | C7H8O | 801 |
| 3 | 17.17 | Eugenol | 61.81 | 164 | C10H12O2 | 958 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, H.M.A.; Alnasser, S.M.; Abd-Elhafeez, H.H.; Alotaibi, M.; Batiha, G.E.-S.; Younis, W. Detection of β-Lactamase Resistance and Biofilm Genes in Pseudomonas Species Isolated from Chickens. Microorganisms 2022, 10, 1975. https://doi.org/10.3390/microorganisms10101975
Mohamed HMA, Alnasser SM, Abd-Elhafeez HH, Alotaibi M, Batiha GE-S, Younis W. Detection of β-Lactamase Resistance and Biofilm Genes in Pseudomonas Species Isolated from Chickens. Microorganisms. 2022; 10(10):1975. https://doi.org/10.3390/microorganisms10101975
Chicago/Turabian StyleMohamed, Hams M. A., Sulaiman Mohammed Alnasser, Hanan H. Abd-Elhafeez, Meshal Alotaibi, Gaber El-Saber Batiha, and Waleed Younis. 2022. "Detection of β-Lactamase Resistance and Biofilm Genes in Pseudomonas Species Isolated from Chickens" Microorganisms 10, no. 10: 1975. https://doi.org/10.3390/microorganisms10101975
APA StyleMohamed, H. M. A., Alnasser, S. M., Abd-Elhafeez, H. H., Alotaibi, M., Batiha, G. E. -S., & Younis, W. (2022). Detection of β-Lactamase Resistance and Biofilm Genes in Pseudomonas Species Isolated from Chickens. Microorganisms, 10(10), 1975. https://doi.org/10.3390/microorganisms10101975