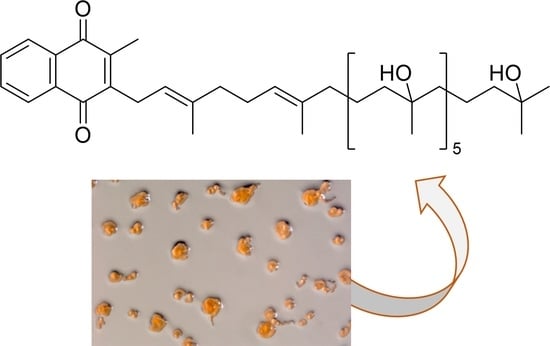
Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of Myxobacteria
2.2. Extraction Procedure
2.3. UPLC-hrMS Analysis
2.4. Purification of Myxoquinones
2.5. NMR Measurements
2.6. Genome Sequencing
3. Results
3.1. Classification of the Producer Organisms
3.2. Identification, Isolation, and Bioactivity Evaluation of Four New Myxoquinones
3.3. Full Structure Elucidation
3.4. Distribution of the Myxoquinones among Myxobacteria
4. Discussion
4.1. Biosynthetic Origin of the Myxoquinones and Biosynthesis Proposal
4.2. On the Possible Biological Role of Myxoquinones
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cenci, U.; Qiu, H.; Pillonel, T.; Cardol, P.; Remacle, C.; Colleoni, C.; Kadouche, D.; Chabi, M.; Greub, G.; Bhattacharya, D.; et al. Host-pathogen biotic interactions shaped vitamin K metabolism in Archaeplastida. Sci. Rep. 2018, 8, 15243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almquist, H.J.; Stokstad, E. Hemorrhagic Chick Disease of Dietary Origin. J. Biol. Chem. 1935, 111, 105–113. [Google Scholar] [CrossRef]
- MacMillan, F.; Hanley, J.; van der Weerd, L.; Knüpling, M.; Un, S.; Rutherford, A.W. Orientation of the phylloquinone electron acceptor anion radical in photosystem I. Biochemistry 1997, 36, 9297–9303. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Plastoquinone in and Beyond Photosynthesis. Trends Plant Sci. 2020, 25, 1252–1265. [Google Scholar] [CrossRef] [PubMed]
- Pralon, T.; Shanmugabalaji, V.; Longoni, P.; Glauser, G.; Ksas, B.; Collombat, J.; Desmeules, S.; Havaux, M.; Finazzi, G.; Kessler, F. Plastoquinone homoeostasis by Arabidopsis proton gradient regulation 6 is essential for photosynthetic efficiency. Commun. Biol. 2019, 2, 220. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.M.; Narasimhulu, K.V.; Samoilova, R.I.; Gennis, R.B.; Dikanov, S.A. Characterization of the semiquinone radical stabilized by the cytochrome aa3-600 menaquinol oxidase of Bacillus subtilis. J. Biol. Chem. 2010, 285, 18241–18251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebedev, A.V.; Ivanova, M.V.; Ruuge, E.K. How do calcium ions induce free radical oxidation of hydroxy-1,4-naphthoquinone? Ca2+ stabilizes the naphthosemiquinone anion-radical of echinochrome A. Arch. Biochem. Biophys. 2003, 413, 191–198. [Google Scholar] [CrossRef]
- Palaniappan, C.; Sharma, V.; Hudspeth, M.E.; Meganathan, R. Menaquinone (vitamin K2) biosynthesis: Evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J. Bacteriol. 1992, 174, 8111–8118. [Google Scholar] [CrossRef] [Green Version]
- Bentley, R.; Meganathan, R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 1982, 46, 241–280. [Google Scholar] [CrossRef]
- Collins, M.D.; Goodfellow, M.; Minnikin, D.E.; Alderson, G. Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J. Appl. Bacteriol. 1985, 58, 77–86. [Google Scholar] [CrossRef]
- Collins, M.D.; Howarth, O.W.; Grund, E.; Kroppenstedt, R.M. Isolation and structural determination of new members of the vitamin K 2 series in Nocardia brasiliensis. FEMS Microbiol. Lett. 1987, 41, 35–39. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, L.; Liu, C.; Sun, Y.; Zhang, D. New aspects of microbial vitamin K2 production by expanding the product spectrum. Microb. Cell Factories 2021, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Panter, F.; Krug, D.; Müller, R. Novel Methoxymethacrylate Natural Products Uncovered by Statistics-Based Mining of the Myxococcus fulvus Secondary Metabolome. ACS Chem. Biol. 2019, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9, 803. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; La Clair, J.J.; Müller, R. Future Directions of Marine Myxobacterial Natural Product Discovery Inferred from Metagenomics. Mar. Drugs 2018, 16, 303. [Google Scholar] [CrossRef] [Green Version]
- Mohr, K.I.; Wolf, C.; Nübel, U.; Szafrańska, A.K.; Steglich, M.; Hennessen, F.; Gemperlein, K.; Kämpfer, P.; Martin, K.; Müller, R.; et al. A polyphasic approach leads to seven new species of the cellulose-decomposing genus Sorangium, Sorangium ambruticinum sp. nov., Sorangium arenae sp. nov., Sorangium bulgaricum sp. nov., Sorangium dawidii sp. nov., Sorangium kenyense sp. nov., Sorangium orientale sp. nov. and Sorangium reichenbachii sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 3576–3586. [Google Scholar] [CrossRef]
- Livingstone, P.G.; Ingleby, O.; Girdwood, S.; Cookson, A.R.; Morphew, R.M.; Whitworth, D.E. Predatory organisms with untapped biosynthetic potential. A description of eight novel Corallococcus species: Corallococcus aberystwythiensis sp. nov., Corallococcus carmarthensis sp. nov., Corallococcus exercitus sp. nov., Corallococcus interemptor sp. nov., Corallococcus llansteffanensis sp. nov., Corallococcus praedator sp. nov., Corallococcus sicarius sp. nov., and Corallococcus terminator sp. nov. Appl. Environ. Microbiol. 2019, 86, e01931-19. [Google Scholar] [CrossRef]
- Chambers, J.; Sparks, N.; Sydney, N.; Livingstone, P.G.; Cookson, A.R.; Whitworth, D.E. Comparative genomics and pan-genomics of the Myxococcaceae, including a description of five novel species: Myxococcus eversor sp. nov., Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis sp. nov., Myxococcus vastator sp. nov., Pyxidicoccus caerfyrddinensis sp. nov. and Pyxidicoccus trucidator sp. nov. Genome Biol. Evol. 2020, 12, 2289–2302. [Google Scholar] [CrossRef]
- Taber, H.W.; Dellers, E.A.; Lombardo, L.R. Menaquinone biosynthesis in Bacillus subtilis: Isolation of men mutants and evidence for clustering of men genes. J. Bacteriol. 1981, 145, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Meganathan, R.; Kwon, O. Biosynthesis of Menaquinone (Vitamin K2) and Ubiquinone (Coenzyme Q). EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [Green Version]
- Daruwala, R.; Kwon, O.; Meganathan, R.; Hudspeth, M.E. A new isochorismate synthase specifically involved in menaquinone (vitamin K2) biosynthesis encoded by the menF gene. FEMS Microbiol. Lett. 1996, 140, 159–163. [Google Scholar] [CrossRef]
- Dawson, A.; Fyfe, P.K.; Hunter, W.N. Specificity and reactivity in menaquinone biosynthesis: The structure of Escherichia coli MenD (2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase). J. Mol. Biol. 2008, 384, 1353–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.; Meganathan, R.; Hudspeth, M.E. Menaquinone (vitamin K2) biosynthesis: Cloning, nucleotide sequence, and expression of the menC gene from Escherichia coli. J. Bacteriol. 1993, 175, 4917–4921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Zhang, H.; Tonge, P.J.; Tan, D.S. Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis. Bioorg. Med. Chem. Lett. 2008, 18, 5963–5966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W. Bringing Bioactive Compounds into Membranes: The UbiA Superfamily of Intramembrane Aromatic Prenyltransferases. Trends Biochem. Sci. 2016, 41, 356–370. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, A.; Schöttler, M.A.; Bréhélin, C.; Kessler, F.; Bock, R.; Cahoon, E.B.; Dörmann, P. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 2006, 281, 40461–40472. [Google Scholar] [CrossRef] [Green Version]
- Kunze, B.; Kemmer, T.; Höfle, G.; Reichenbach, H. Stigmatellin, a new antibiotic from Stigmatella aurantiaca (Myxobacterales). I. Production, physico-chemical and biological properties. J. Antibiot. 1984, 37, 454–461. [Google Scholar] [CrossRef] [Green Version]
- Gerth, K.; Irschik, H.; Reichenbach, H.; Trowitzsch, W. Myxothiazol, an antibiotic from Myxococcus fulvus (myxobacterales). I. Cultivation, isolation, physico-chemical and biological properties. J. Antibiot. 1980, 33, 1474–1479. [Google Scholar] [CrossRef] [Green Version]
- Weinig, S.; Hecht, H.-J.; Mahmud, T.; Müller, R. Melithiazol biosynthesis: Further insights into myxobacterial PKS/NRPS systems and evidence for a new subclass of methyl transferases. Chem. Biol. 2003, 10, 939–952. [Google Scholar] [CrossRef] [Green Version]
- Gröber, U.; Reichrath, J.; Holick, M.F.; Kisters, K. Vitamin K: An old vitamin in a new perspective. Dermatoendocrinology 2014, 6, e968490. [Google Scholar] [CrossRef] [Green Version]
- Ernster, L.; Dallner, G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta Mol. Basis Dis. 1995, 1271, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.D.; Jones, D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 1981, 45, 316–354. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.W.A.; Hellingwerf, K.J. All Three Endogenous Quinone Species of Escherichia coli Are Involved in Controlling the Activity of the Aerobic/Anaerobic Response Regulator ArcA. Front. Microbiol. 2016, 7, 1339. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, M. Vitamin K2 Biosynthesis: Drug Targets for New Antibacterials. In Vitamin K2—Vital for Health and Wellbeing; Gordeladze, J.O., Ed.; InTech: London, UK, 2017; ISBN 978-953-51-3019-2. [Google Scholar]
- Shearer, M.J.; Newman, P. Recent trends in the metabolism and cell biology of vitamin K with special reference to vitamin K cycling and MK-4 biosynthesis. J. Lipid Res. 2014, 55, 345–362. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panter, F.; Popoff, A.; Garcia, R.; Krug, D.; Müller, R. Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones. Microorganisms 2022, 10, 534. https://doi.org/10.3390/microorganisms10030534
Panter F, Popoff A, Garcia R, Krug D, Müller R. Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones. Microorganisms. 2022; 10(3):534. https://doi.org/10.3390/microorganisms10030534
Chicago/Turabian StylePanter, Fabian, Alexander Popoff, Ronald Garcia, Daniel Krug, and Rolf Müller. 2022. "Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones" Microorganisms 10, no. 3: 534. https://doi.org/10.3390/microorganisms10030534
APA StylePanter, F., Popoff, A., Garcia, R., Krug, D., & Müller, R. (2022). Myxobacteria of the Cystobacterineae Suborder Are Producers of New Vitamin K2 Derived Myxoquinones. Microorganisms, 10(3), 534. https://doi.org/10.3390/microorganisms10030534