Metabolism of Aldoximes and Nitriles in Plant-Associated Bacteria and Its Potential in Plant-Bacteria Interactions
Abstract
:1. Introduction
2. Substrate Specificity
2.1. Aldoxime Dehydratases
2.2. Nitrile-Converting Enzymes
3. Taxonomic and Ecological Distribution
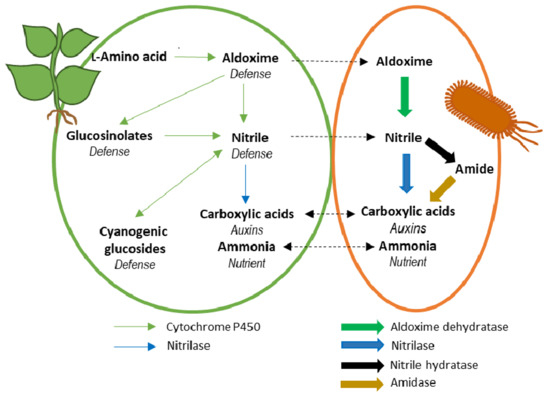
4. Potentiality of the Aldoxime-Nitrile Pathway in Plant-Associated Bacteria
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sørensen, M.; Neilson, E.H.J.; Møller, B.L. Oximes: Unrecognized chameleons in general and specialized plant metabolism. Mol. Plant 2018, 11, 95–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howden, A.J.; Preston, G.M. Nitrilase enzymes and their role in plant-microbe interactions. Microb. Biotechnol. 2009, 2, 441–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowski, M. Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 2008, 69, 2655–2667. [Google Scholar] [CrossRef]
- Kato, Y.; Tsuda, T.; Asano, Y. Purification and partial characterization of N-hydroxy-l-phenylalanine decarboxylase/oxidase from Bacillus sp. strain OxB-1, an enzyme involved in aldoxime biosynthesis in the “aldoxime-nitrile pathway”. Biochim. Biophys. Acta 2007, 1774, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Betke, T.; Higuchi, J.; Rommelmann, P.; Oike, K.; Nomura, T.; Kato, Y.; Asano, Y.; Gröger, H. Biocatalytic synthesis of nitriles through dehydration of aldoximes: The substrate scope of aldoxime dehydratases. ChemBioChem 2018, 19, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L. Nitrile metabolism in fungi: A review of its key enzymes nitrilases with focus on their biotechnological impact. Fungal Biol. Rev. 2019, 33, 149–157. [Google Scholar] [CrossRef]
- Shen, J.-D.; Cai, X.; Liu, Z.-Q.; Zheng, Y.-G. Nitrilase: A promising biocatalyst in industrial applications for green chemistry. Crit. Rev. Biotechnol. 2020, 41, 72–93. [Google Scholar] [CrossRef]
- Stolz, A.; Eppinger, E.; Sosedov, O.; Kiziak, C. Comparative analysis of the conversion of mandelonitrile and 2-phenylpropionitrile by a large set of variants generated from a nitrilase originating from Pseudomonas fluorescens EBC191. Molecules 2019, 24, 4232. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, T.C.; Kumar, V.; Kumar, V. Enzymes of aldoxime-nitrile pathway for organic synthesis. Rev. Environ. Sci. Bio-Technol. 2018, 17, 229–239. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda Mdel, C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, S. Conversion of 3-indolacetaldoxime to 3-indoleacetonitrile by plants. Arch. Biochem. Biophys. 1963, 100, 557–558. [Google Scholar] [CrossRef]
- Kato, Y.; Ooi, R.; Asano, Y. Isolation and characterization of a bacterium possessing a novel aldoxime-dehydration activity and nitrile-degrading enzymes. Arch. Microbiol. 1998, 170, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Nakamura, K.; Sakiyama, H.; Mayhew, S.G.; Asano, Y. Novel heme-containing lyase, phenylacetaldoxime dehydratase from Bacillus sp. strain OxB-1: Purification, characterization, and molecular cloning of the gene. Biochemistry 2000, 39, 800–809. [Google Scholar] [CrossRef]
- Oinuma, K.; Hashimoto, Y.; Konishi, K.; Goda, M.; Noguchi, T.; Higashibata, H.; Kobayashi, M. Novel aldoxime dehydratase involved in carbon-nitrogen triple bond synthesis of Pseudomonas chlororaphis B23. Sequencing, gene expression, purification, and characterization. J. Biol. Chem. 2003, 278, 29600–29608. [Google Scholar] [CrossRef] [Green Version]
- Martínková, L.; Rucká, L.; Nešvera, J.; Pátek, M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J. Microbiol. Biotechnol. 2017, 33, 11. [Google Scholar] [CrossRef]
- Asano, Y.; Tani, Y.; Yamada, H. A new enzyme nitrile hydratase which degrades acetonitrile in combination with amidase. Agr. Biol. Chem. 1980, 44, 2251–2252. [Google Scholar] [CrossRef] [Green Version]
- Hinzmann, A.; Betke, T.; Asano, Y.; Gröger, H. Synthetic processes toward nitriles without the use of cyanide: A biocatalytic concept based on dehydration of aldoximes in water. Chem. Eur. J. 2021, 27, 5313–5321. [Google Scholar] [CrossRef]
- Pedras, M.S.; Minic, Z.; Thongbam, P.D.; Bhaskar, V.; Montaut, S. Indolyl-3-acetaldoxime dehydratase from the phytopathogenic fungus Sclerotinia sclerotiorum: Purification, characterization, and substrate specificity. Phytochemistry 2010, 71, 1952–1962. [Google Scholar] [CrossRef]
- Kato, Y.; Asano, Y. Purification and characterization of aldoxime dehydratase of the head blight fungus, Fusarium graminearum. Biosci. Biotechnol. Biochem. 2005, 69, 2254–2257. [Google Scholar] [CrossRef] [Green Version]
- Kato, Y.; Asano, Y. Molecular and enzymatic analysis of the “aldoxime-nitrile pathway” in the glutaronitrile degrader Pseudomonas sp. K-9. Appl. Microbiol. Biotechnol. 2006, 70, 92–101. [Google Scholar] [CrossRef]
- Xie, S.X.; Kato, Y.; Komeda, H.; Yoshida, S.; Asano, Y. A gene cluster responsible for alkylaldoxime metabolism coexisting with nitrile hydratase and amidase in Rhodococcus globerulus A-4. Biochemistry 2003, 42, 12056–12066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Yoshida, S.; Xie, S.-X.; Asano, Y. Aldoxime dehydratase co-existing with nitrile hydratase and amidase in the iron-type nitrile hydratase-producer Rhodococcus sp. N-771. J. Biosci. Bioeng. 2004, 97, 250–259. [Google Scholar] [CrossRef]
- Rädisch, R.; Chmátal, M.; Rucká, L.; Novotný, P.; Petrásková, L.; Halada, P.; Kotík, M.; Pátek, M.; Martínková, L. Overproduction and characterization of the first enzyme of a new aldoxime dehydratase family in Bradyrhizobium sp. Int. J. Biol. Macromol. 2018, 115, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Asano, Y. High-level expression of a novel FMN-dependent heme-containing lyase, phenylacetaldoxime dehydratase of Bacillus sp. strain OxB-1, in heterologous hosts. Protein Expres. Purif. 2003, 28, 131–139. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov (accessed on 2 February 2022).
- Knoch, E.; Motawie, M.S.; Olsen, C.E.; Møller, B.L.; Lyngkjaer, M.F. Biosynthesis of the leucine derived alpha-, beta- and gamma-hydroxynitrile glucosides in barley (Hordeum vulgare L.). Plant. J. 2016, 88, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Tsuda, T.; Asano, Y. Nitrile hydratase involved in aldoxime metabolism from Rhodococcus sp. strain YH3-3 purification and characterization. Eur. J. Biochem. 1999, 263, 662–670. [Google Scholar] [CrossRef] [Green Version]
- Nagasawa, T.; Nanba, H.; Ryuno, K.; Takeuchi, K.; Yamada, H. Nitrile hydratase of Pseudomonas chlororaphis B23. Eur. J. Biochem. 1987, 162, 691–698. [Google Scholar] [CrossRef]
- Nagamune, T.; Kurata, H.; Hirata, M.; Honda, J.; Koike, H.; Ikeuchi, M.; Inoue, Y.; Hirata, A.; Endo, I. Purification of inactivated photoresponsive nitrile hydratase. Biochem. Biophys. Res. Commun. 1990, 168, 437–442. [Google Scholar] [CrossRef]
- Nojiri, M.; Nakayama, H.; Odaka, M.; Yohda, M.; Takio, K.; Endo, I. Cobalt-substituted Fe-type nitrile hydratase of Rhodococcus sp. N-771. FEBS Lett. 2000, 465, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Duca, D.; Rose, D.R.; Glick, B.R. Characterization of a nitrilase and a nitrile hydratase from Pseudomonas sp. strain UW4 that converts indole-3-acetonitrile to indole-3-acetic acid. Appl. Environ. Microbiol. 2014, 80, 4640–4649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, D.E.; Chaplin, J.A.; DeSantis, G.; Podar, M.; Madden, M.; Chi, E.; Richardson, T.; Milan, A.; Miller, M.; Weiner, D.P.; et al. Exploring nitrilase sequence space for enantioselective catalysis. Appl. Environ. Microbiol. 2004, 70, 2429–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Mukherjee, C.; Yang, Y.; Rios, B.E.; Gallagher, D.T.; Smith, N.N.; Biehl, E.R.; Hua, L. A new nitrilase from Bradyrhizobium japonicum USDA 110. Gene cloning, biochemical characterization and substrate specificity. J. Biotechnol. 2008, 133, 327–333. [Google Scholar] [CrossRef]
- Kato, Y.; Ooi, R.; Asano, Y. Distribution of aldoxime dehydratase in microorganisms. Appl. Environ. Microbiol. 2000, 66, 2290–2296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Yoshida, S.; Asano, Y. Polymerase chain reaction for identification of aldoxime dehydratase in aldoxime- or nitrile-degrading microorganisms. FEMS Microbiol. Lett. 2005, 246, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Mishra, V.; Rau, N.; Sharma, R.S. Increased iron-stress resilience of maize through inoculation of siderophore-producing Arthrobacter globiformis from mine. J. Basic. Microbiol. 2016, 56, 719–735. [Google Scholar] [CrossRef]
- Worsley, S.F.; Newitt, J.; Rassbach, J.; Batey, S.F.D.; Holmes, N.A.; Murrell, J.C.; Wilkinson, B.; Hutchings, M.I. Streptomyces endophytes promote host health and enhance growth across plant species. Appl. Environ. Microbiol. 2020, 86, e01053-20. [Google Scholar] [CrossRef]
- Vick, S.H.W.; Fabian, B.K.; Dawson, C.J.; Foster, C.; Asher, A.; Hassan, K.A.; Midgley, D.J.; Paulsen, I.T.; Tetu, S.G. Delving into defence: Identifying the Pseudomonas protegens Pf-5 gene suite involved in defence against secreted products of fungal, oomycete and bacterial rhizosphere competitors. Microb. Genom. 2021, 7, 671. [Google Scholar] [CrossRef]
- Gonzalez-Benitez, N.; Martin-Rodriguez, I.; Cuesta, I.; Arrayas, M.; White, J.F.; Molina, M.C. Endophytic microbes are tools to increase tolerance in Jasione plants against arsenic stress. Front. Microbiol. 2021, 12, 664271. [Google Scholar] [CrossRef]
- Yandigeri, M.S.; Meena, K.K.; Singh, D.; Malviya, N.; Singh, D.P.; Solanki, M.K.; Yadav, A.K.; Arora, D.K. Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 2012, 68, 411–420. [Google Scholar] [CrossRef]
- Song, L.; Wang, M.Z.; Shi, J.J.; Xue, Z.Q.; Wang, M.X.; Qian, S.J. High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism. Biochem. Biophys. Res. Commun. 2007, 362, 319–324. [Google Scholar] [CrossRef]
- VanInsberghe, D.; Maas, K.R.; Cardenas, E.; Strachan, C.R.; Hallam, S.J.; Mohn, W.W. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J. 2015, 9, 2435–2441. [Google Scholar] [CrossRef] [Green Version]
- Jones, F.P.; Clark, I.M.; King, R.; Shaw, L.J.; Woodward, M.J.; Hirsch, P.R. Novel European free-living, non-diazotrophic Bradyrhizobium isolates from contrasting soils that lack nodulation and nitrogen fixation genes—A genome comparison. Sci. Rep. 2016, 6, 25858. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Babalola, O.O.; Glick, B.R. Plant growth-promoting root-colonizing bacterial endophytes. Rhizosphere 2021, 20, 100433. [Google Scholar] [CrossRef]
- Han, J.I.; Choi, H.K.; Lee, S.W.; Orwin, P.M.; Kim, J.; Laroe, S.L.; Kim, T.G.; O’Neil, J.; Leadbetter, J.R.; Lee, S.Y.; et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011, 193, 1183–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, M.; Archana, G. Phenotypic display of plant growth-promoting traits in individual strains and multispecies consortia of plant growth promoting rhizobacteria and rhizobia under salinity stress. Rhizosphere 2021, 20, 443. [Google Scholar] [CrossRef]
- Guo, H.; Glaeser, S.P.; Alabid, I.; Imani, J.; Haghighi, H.; Kämpfer, P.; Kogel, K.H. The abundance of endofungal bacterium Rhizobium radiobacter (syn. Agrobacterium tumefaciens) increases in its fungal host Piriformospora indica during the tripartite sebacinalean symbiosis with higher plants. Front. Microbiol. 2017, 8, 629. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, M.I.P.; Nascimento, R.d.C.; Rodrigues, D.R.; Escobar, I.E.C.; Fraiz, A.C.R.; de Souza, A.P.; de Freitas, A.D.S.; Nóbrega, R.S.A.; Fernandes-Júnior, P.I. Maize growth and yield promoting endophytes isolated into a legume root nodule by a cross-over approach. Rhizosphere 2020, 15, 100211. [Google Scholar] [CrossRef]
- Thuku, R.N.; Brady, D.; Benedik, M.J.; Sewell, B.T. Microbial nitrilases: Versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 2009, 106, 703–727. [Google Scholar] [CrossRef] [Green Version]
- Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome mining and evaluation of the biocontrol potential of Pseudomonas fluorescens BRZ63, a new endophyte of oilseed rape (Brassica napus L.) against fungal pathogens. Int. J. Mol. Sci. 2020, 21, 8740. [Google Scholar] [CrossRef]
- Jiang, F.; Chen, L.; Belimov, A.A.; Shaposhnikov, A.I.; Gong, F.; Meng, X.; Hartung, W.; Jeschke, D.W.; Davies, W.J.; Dodd, I.C. Multiple impacts of the plant growth-promoting rhizobacterium Variovorax paradoxus 5C-2 on nutrient and ABA relations of Pisum sativum. J. Exp. Bot. 2012, 63, 6421–6430. [Google Scholar] [CrossRef] [PubMed]
- Omura, H.; Kuroda, M.; Kobayashi, M.; Shimizu, S.; Yoshida, T.; Nagasawa, T. Purification, characterization and gene cloning of thermostable O-acetyl-L-serine sulfhydrylase forming beta-cyano-L-alanine. J. Biosci. Bioeng. 2003, 95, 470–475. [Google Scholar] [CrossRef]
- Howden, A.J.M.; Jill Harrison, C.; Preston, G.M. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 2009, 57, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Rucká, L.; Kulik, N.; Novotný, P.; Sedova, A.; Petrásková, L.; Příhodová, R.; Křístková, B.; Halada, P.; Pátek, M.; Martínková, L. Plant nitrilase homologues in fungi: Phylogenetic and functional analysis with focus on nitrilases in Trametes versicolor and Agaricus bisporus. Molecules 2020, 25, 3861. [Google Scholar] [CrossRef] [PubMed]
- Bruto, M.; Prigent-Combaret, C.; Luis, P.; Moënne-Loccoz, Y.; Muller, D. Frequent, independent transfers of a catabolic gene from bacteria to contrasted filamentous eukaryotes. Proc. R. Soc. B 2014, 281, 20140848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casanova-Sáez, R.; Voß, U. Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 2019, 24, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef] [Green Version]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [Green Version]
- Bogner, C.W.; Kamdem, R.S.; Sichtermann, G.; Matthäus, C.; Holscher, D.; Popp, J.; Proksch, P.; Grundler, F.M.; Schouten, A. Bioactive secondary metabolites with multiple activities from a fungal endophyte. Microb. Biotechnol. 2017, 10, 175–188. [Google Scholar] [CrossRef]
- Hernández-León, R.; Rojas-Solís, D.; Contreras-Pérez, M.; Orozco-Mosqueda, M.d.C.; Macías-Rodríguez, L.I.; Reyes-de la Cruz, H.; Valencia-Cantero, E.; Santoyo, G. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 2015, 81, 83–92. [Google Scholar] [CrossRef]
- Kiziak, C.; Conradt, D.; Stolz, A.; Mattes, R.; Klein, J. Nitrilase from Pseudomonas fluorescens EBC191: Cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 2005, 151, 3639–3648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdellatif, L.; Ben-Mahmoud, O.M.; Yang, C.; Hanson, K.G.; Gan, Y.; Hamel, C. The H2-oxidizing rhizobacteria associated with field-grown lentil promote the growth of lentil inoculated with hup+ Rhizobium through multiple modes of action. J. Plant Growth Regul. 2016, 36, 348–361. [Google Scholar] [CrossRef]
- Zhao, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Wu, G.H.; Tian, C.Y. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 2016, 73, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.J.; Yin, B.; Wang, C.; Jiang, S.Q.; Wang, H.L.; Yuan, Y.A.; Wei, D.Z. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803. J. Struct. Biol. 2014, 188, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Engels, D.; Bürger, S.; Kiziak, C.; Mattes, R.; Stolz, A. Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl. Environ. Microbiol. 2003, 69, 4359–4366. [Google Scholar] [CrossRef] [Green Version]
- Rucká, L.; Chmátal, M.; Kulik, N.; Petrásková, L.; Pelantová, H.; Novotný, P.; Příhodová, R.; Pátek, M.; Martínková, L. Genetic and functional diversity of nitrilases in Agaricomycotina. Int. J. Mol. Sci. 2019, 20, 5990. [Google Scholar] [CrossRef] [Green Version]
- Savory, E.A.; Fuller, S.L.; Weisberg, A.J.; Thomas, W.J.; Gordon, M.I.; Stevens, D.M.; Creason, A.L.; Belcher, M.S.; Serdani, M.; Wiseman, M.S.; et al. Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management. Elife 2017, 6, e30925. [Google Scholar] [CrossRef] [Green Version]
- Savory, E.A.; Weisberg, A.J.; Stevens, D.M.; Creason, A.L.; Fuller, S.L.; Pearce, E.M.; Chang, J.H. Phytopathogenic Rhodococcus have diverse plasmids with few conserved virulence functions. Front. Microbiol. 2020, 11, 1022. [Google Scholar] [CrossRef]
- Vandeputte, O.; Oden, S.; Mol, A.; Vereecke, D.; Goethals, K.; El Jaziri, M.; Prinsen, E. Biosynthesis of auxin by the gram-positive phytopathogen Rhodococcus fascians is controlled by compounds specific to infected plant tissues. Appl. Environ. Microbiol. 2005, 71, 1169–1177. [Google Scholar] [CrossRef] [Green Version]







| OxdB (BAA90461.1) | OxdRG (BAC99076.1) 1 | OxdK (BAD98528.1) 2 | OxdBr1 (WP_044589203) | |
|---|---|---|---|---|
| (Bacillus sp. OxB-1) [25] | (Rhododoccus globerulus A-4 [22] | (Pseudomonas sp. K-9) [21] | (Bradyrhizobium sp. LTSPM299) [24] | |
| OxdB | − | 32 (92) | 32 (93) | 32 (93) |
| OxdRG | − | − | 76 (99) | 47 (98) |
| OxdK | − | − | − | 47 (99) |
| Enzyme 1 | Substrate 2 | Relative Activity (%) | Vmax [U/mg] | KM [mM] | Vmax/KM [U/mg/mM] |
|---|---|---|---|---|---|
| OxdB [14] | IAOx | 18 | 5.42 | 2.40 | 2.26 |
| iVaOx | 20 | 7.72 | 3.58 | 2.16 | |
| Z-PAOx | 100 | 19.5 | 0.872 | 22.4 | |
| Z-PPOx | 63 | 14.3 | 1.36 | 10.5 | |
| OxdK [21] | IAOx | 1 | n.d. | n.d. | n.d. |
| iBuOx | 14 | 5.87 | 0.538 | 10.9 | |
| iVaOx | 100 | 35.1 | 1.33 | 26.4 | |
| Z-PAOx | 7 | 2.61 | 0.991 | 2.63 | |
| Z-PPOx | 31 | 12.1 | 0.975 | 12.4 | |
| OxdRG [22] | IAOx | 11 | 0.281 | 3.91 | 0.072 |
| iBuOx | 39 | 0.041 | 5.54 | 0.007 | |
| iVaOx | 87 | 0.239 | 3.97 | 0.060 | |
| Z-PAOx | 40 | 0.140 | 1.40 | 0.100 | |
| Z-PPOx | 100 | 0.392 | 2.31 | 0.170 |
| Phylum | Genus | Number of Sequences 1 | Number of Active Strains 2 | ||
|---|---|---|---|---|---|
| Oxd | NLase | NHase | |||
| Actinobacteria | Arthrobacter | 5 | 16 | 5 | n.d. |
| Cellulomonas | - | 10 | - | n.d. | |
| Corynebacterium | 3 | 3 | - | 2 | |
| Brevibacterium | 1 | 41 | 1 | n.d. | |
| Gordonia | 4 | 6 | 13 | n.d. | |
| Microbacterium | 5 | 27 | 9 | 1 | |
| Nitriliruptor | 2 | 3 | 1 | n.a. | |
| Micrococcus | - | - | - | 1 | |
| Nocardia | 1 | 36 | 7 | 2 | |
| Rhodococcus | 142 | 190 | 107 | 14 | |
| Streptomyces | 8 | 157 | 252 | n.d. | |
| Firmicutes | Bacillus | 2 | 15 | 2 | 2 |
| Brevibacillus | 2 | 66 | - | n.a. | |
| Cytobacillus | 1 | 1 | - | n.a. | |
| Geomicrobium | 1 | 1 | - | n.a. | |
| Paenibacillus | 1 | 99 | 13 | n.a. | |
| Siminovitchia | 3 | 14 | - | n.a. | |
| Proteobacteria | Afipia | 2 | - | 14 | n.a. |
| Agrobacterium | 9 | 99 | 43 | n.d. | |
| Bradyrhizobium | 248 | 101 | 298 | n.a. | |
| Erwinia | 1 | 19 | 5 | n.d. | |
| Gibbsiella | 1 | 10 | 2 | n.a. | |
| Herbaspirillum | 1 | 45 | 9 | n.a. | |
| Klebsiella | - | 17 | 66 | n.d. | |
| Novosphingobium | 1 | 68 | 1 | n.a. | |
| Paraburkholderia | 1 | 336 | 139 | n.a. | |
| Proteus | - | 1 | 2 | n.d. | |
| Pseudomonas | 48 | 261 | 50 | 2 | |
| Pseudomaricurvus | 1 | - | - | n.a. | |
| Rhizobium | 7 | 270 | 600 | n.a. | |
| Serratia | - | 4 | 11 | n.d. | |
| Variovorax | 73 | 156 | 92 | n.d. | |
| Xanthomonas | 2 | 1 | 2 | n.d. | |
| Bacteroidetes 3 | Flavobacterium | - | 172 | 4 | n.d. |
| Maribacter | 1 | 58 | - | n.a. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rädisch, R.; Pátek, M.; Křístková, B.; Winkler, M.; Křen, V.; Martínková, L. Metabolism of Aldoximes and Nitriles in Plant-Associated Bacteria and Its Potential in Plant-Bacteria Interactions. Microorganisms 2022, 10, 549. https://doi.org/10.3390/microorganisms10030549
Rädisch R, Pátek M, Křístková B, Winkler M, Křen V, Martínková L. Metabolism of Aldoximes and Nitriles in Plant-Associated Bacteria and Its Potential in Plant-Bacteria Interactions. Microorganisms. 2022; 10(3):549. https://doi.org/10.3390/microorganisms10030549
Chicago/Turabian StyleRädisch, Robert, Miroslav Pátek, Barbora Křístková, Margit Winkler, Vladimír Křen, and Ludmila Martínková. 2022. "Metabolism of Aldoximes and Nitriles in Plant-Associated Bacteria and Its Potential in Plant-Bacteria Interactions" Microorganisms 10, no. 3: 549. https://doi.org/10.3390/microorganisms10030549
APA StyleRädisch, R., Pátek, M., Křístková, B., Winkler, M., Křen, V., & Martínková, L. (2022). Metabolism of Aldoximes and Nitriles in Plant-Associated Bacteria and Its Potential in Plant-Bacteria Interactions. Microorganisms, 10(3), 549. https://doi.org/10.3390/microorganisms10030549