Amino Acid-Derived Bacterial Metabolites in the Colorectal Luminal Fluid: Effects on Microbial Communication, Metabolism, Physiology, and Growth
Abstract
:1. Introduction
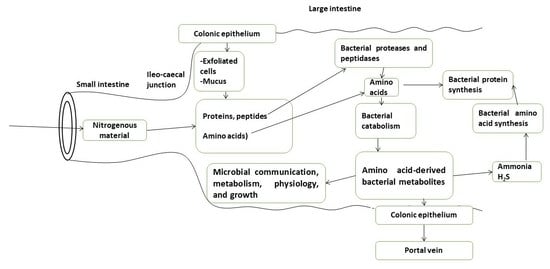
2. Bacteria in the Large Intestinal Luminal Fluid Release Amino Acids from Undigested Proteins and Used Them for Protein Synthesis, Energy Production, and Other Catabolic Pathways Which Release Various Bacterial Metabolites
3. Amino Acid-Derived Bacterial Metabolites Are Involved in the Biology of Intestinal Microbes
3.1. Lactate, Formate, Succinate and Oxaloacetate
3.2. p-Cresol
3.3. Indole
3.4. Skatole
3.5. Hydrogen Sulfide
3.6. Polyamines
3.7. Gamma-Amino Butyric Acid, Norepinephrine, and Serotonin
3.8. 4-Hydroxyphenylacetate
4. Conclusions and Perspectives
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bandyopadhyay, S.; Kashyap, S.; Calvez, J.; Devi, S.; Azzout-Marniche, D.; Tomé, D.; Kurpad, A.V.; Gaudichon, C. Evaluation of protein quality in humans and insights on stable isotope approaches to measure digestibility—A Review. Adv. Nutr. 2022, 13, 1131–1143. [Google Scholar] [CrossRef]
- Lister, N.; Sykes, A.P.; Bailey, P.D.; Boyd, C.A.; Bronk, J.R. Dipeptide transport and hydrolysis in isolated loops of rat small intestine: Effects of stereospecificity. J. Physiol. 1995, 484, 173–182. [Google Scholar] [CrossRef]
- Buddington, R.K.; Elnif, J.; Puchal-Gardiner, A.A.; Sangild, P.T. Intestinal apical amino acid absorption during development of the pig. Am. J. Physiol. 2001, 280, R241–R247. [Google Scholar] [CrossRef]
- Broër, S.; Fairweather, S.J. Amino acid transport across the mammalian intestine. Compr. Physiol. 2018, 9, 343–373. [Google Scholar]
- Gibson, J.A.; Sladen, G.E.; Dawson, A.M. Protein absorption and ammonia production: The effects of dietary protein and removal of the colon. Br. J. Nutr. 1976, 35, 61–65. [Google Scholar] [CrossRef]
- Kramer, P. The effect of varying sodium loads on the ileal excreta of human ileostomized subjects. J. Clin. Investig. 1966, 45, 1710–1718. [Google Scholar] [CrossRef]
- Smiddy, F.G.; Gregory, S.D.; Smith, I.B.; Goligher, J.C. Faecal loss of fluid, electrolytes, and nitrogen in colitis before and after ileostomy. Lancet 1960, 1, 14–19. [Google Scholar] [CrossRef]
- Chacko, A.; Cummings, J.H. Nitrogen losses from the human small bowel: Obligatory losses and the effect of physical form of food. Gut 1988, 29, 809–815. [Google Scholar] [CrossRef]
- Gaudichon, C.; Bos, C.; Morens, C.; Petzke, K.J.; Mariotti, F.; Everwand, J.; Benamouzig, R.; Daré, S.; Tomé, D.; Metges, C.C. Ileal losses of nitrogen and amino acids in humans and their importance to the assessment of amino acid requirements. Gastroenterology 2002, 123, 50–59. [Google Scholar] [CrossRef]
- Webb, K.E., Jr.; Matthews, J.C.; DiRienzo, D.B. Peptide absorption: A review of current concepts and future perspectives. J. Anim. Sci. 1992, 70, 3248–3257. [Google Scholar] [CrossRef]
- Webb, K.E., Jr.; DiRienzo, D.B.; Matthews, J.C. Recent developments in gastrointestinal absorption and tissue utilization of peptides: A review. J. Dairy Sci. 1993, 76, 351–361. [Google Scholar] [CrossRef]
- Dave, L.A.; Montoya, C.A.; Rutherfurd, S.M.; Moughan, P.J. Gastrointestinal endogenous proteins as a source of bioactive peptides--an in silico study. PLoS ONE 2014, 9, e98922. [Google Scholar] [CrossRef]
- Mariotti, F.; Tomé, D.; Mirand, P.P. Converting nitrogen into protein--beyond 6.25 and Jones’ factors. Crit. Rev. Food. Sci. Nutr. 2008, 48, 177–184. [Google Scholar] [CrossRef]
- Dubuisson, C.; Lioret, S.; Touvier, M.; Dufour, A.; Calamassi-Tran, G.; Volatier, J.L.; Lafay, L. Trends in food and nutritional intakes of French adults from 1999 to 2007: Results from the INCA surveys. Br. J. Nutr. 2010, 103, 1035–1048. [Google Scholar] [CrossRef]
- Pasiakos, S.M.; Agarwal, S.; Lieberman, H.R.; Fulgoni, V.L., 3rd. Sources and amounts of animal, dairy, and plant protein intake of US adults in 2007–2010. Nutrients 2015, 7, 7058–7069. [Google Scholar] [CrossRef]
- Kashyap, S.; Shivakumar, N.; Varkey, A.; Duraisamy, R.; Thomas, T.; Preston, T.; Devi, S.; Kurpad, A.V. Ileal digestibility of intrinsically labeled hen’s egg and meat protein determined with the dual stable isotope tracer method in Indian adults. Am. J. Clin. Nutr. 2018, 108, 980–987. [Google Scholar] [CrossRef]
- Bharucha, A.E.; Anderson, B.; Bouchoucha, M. More movement with evaluating colonic transit in humans. Neurogastroenterol. Motil. 2019, 31, e13541. [Google Scholar] [CrossRef]
- Rose, C.; Parker, A.; Jefferson, B.; Cartmell, E. The characterization of feces and urine: A review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1827–1879. [Google Scholar] [CrossRef]
- Raymann, K.; Moeller, A.H.; Goodman, A.L.; Ochman, H. Unexplored archaeal diversity in the great ape Gut Microbiome. mSphere 2017, 2, e00026-17. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The human gut virome Is highly diverse, stable, and individual specific. Cell Host Microbe 2019, 26, 527–541.e5. [Google Scholar] [CrossRef]
- Carding, S.R.; Davis, N.; Hoyles, L. Review article: The human intestine virome in health and disease. Aliment. Pharmacol. Ther. 2017, 46, 800–815. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.J.; Oh, S.; Underhill, D.M. Host-microbe interactions: Commensal fungi in the gut. Curr. Opin. Microbiol. 2017, 40, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.L.; Gilchrist, C.A.; Lynn, T.C.; Petri, W.A., Jr. Parasitic protozoan and interactions with the host intestinal microbiota. Infect. Immunol. 2017, 85, e00101-17. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F. Metabolism of Alimentary Compounds by the Intestinal Microbiota and Health; Springer: Wien, Austria, 2023. [Google Scholar]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Kaman, W.E.; Hays, J.P.; Endtz, H.P.; Bikker, F.J. Bacterial proteases: Targets for diagnostics and therapy. Eur. J. Clin.Microbiol. Infect. Dis. 2014, 33, 1081–1087. [Google Scholar] [CrossRef]
- Cristofori, F.; Francavilla, R.; Capobianco, D.; Dargenio, V.N.; Filardo, S.; Mastromarino, P. Bacterial-based strategies to hydrolyze gluten peptides and protect intestinal mucosa. Front. Immunol. 2020, 11, 567801. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Cummings, J.H. The Colonic flora, fermentation, and large bowel digestive function. In The Large Intestine; Phillips, S.F., Pemberton, J.H., Shorter, R.G., Eds.; Raven Press: New York, NY, USA, 1991; pp. 51–92. [Google Scholar]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef]
- Steiner, H.Y.; Naider, F.; Becker, J.M. The PTR family: A new group of peptide transporters. Mol. Microbiol. 1995, 16, 825–834. [Google Scholar] [CrossRef]
- Davies, J.S.; Currie, M.J.; Wright, J.D.; Newton-Vesty, M.C.; North, R.A.; Mace, P.D.; Allison, J.R.; Dobson, R.C.J. Selective nutrient transport in bacteria: Multicomponent transporter systems reign supreme. Front. Mol. Biosci. 2021, 8, 699222. [Google Scholar] [CrossRef]
- Garai, P.; Chandra, K.; Chakravortty, D. Bacterial peptide transporters: Messengers of nutrition to virulence. Virulence 2017, 8, 297–309. [Google Scholar] [CrossRef]
- Eggeling, L.; Sahm, H. New ubiquitous translocators: Amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 2003, 180, 155–160. [Google Scholar]
- Katsube, S.; Ando, T.; Yoneyama, H. L-Alanine exporter, AlaE, of Escherichia coli functions as a safety valve to enhance survival under feast conditions. Int. J. Mol. Sci. 2019, 20, 4942. [Google Scholar] [CrossRef]
- James, P.S.; Smith, M.V. Methionine transport by pig colonic mucosa measured during early post-natal development. J. Physiol. 1976, 262, 151–168. [Google Scholar] [CrossRef]
- Sepulveda, F.V.; Smith, M.W. Different mechanisms for neutral amino acid uptake by new-born colon. J. Physiol. 1979, 286, 479–490. [Google Scholar] [CrossRef]
- van der Wielen, N.; Moughan, P.J.; Mensink, M. Amino acid absorption in the large intestine of human and porcine models. J. Nutr. 2017, 147, 1493–1498. [Google Scholar] [CrossRef]
- Wuensch, T.; Schulz, S.; Ullrich, S.; Lill, N.; Stelzl, T.; Rubio-Aliaga, I.; Loh, G.; Chamaillard, M.; Haller, D.; Daniel, H. The peptide transporter PEPT1 is expressed in distal colon in rodents and humans and contributes to water absorption. Am. J. Physiol. 2013, 305, G66–G73. [Google Scholar] [CrossRef]
- Stephen, A.M.; Cummings, J.H. The microbial contribution to human fecal mass. J. Med. Microbiol. 1980, 13, 45–56. [Google Scholar] [CrossRef]
- Portune, K.J.; Beaumont, M.; Davila, A.M.; Tomé, D.; Blachier, F.; Sanz, Y. Gut microbiota role in dietary protein metabolism and health-related outcomes: The two sides of the coin. Trends Food. Sci. Technol. 2016, 57, 213–232. [Google Scholar] [CrossRef]
- Smith, A.B.; Jenior, M.L.; Keenan, O.; Hart, J.L.; Specker, J.; Abbas, A.; Rangel, P.C.; Di, C.; Green, J.; Bustin, K.A.; et al. Enterococci enhance Clostridioides difficile pathogenesis. Nature 2022, 611, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ohtani, K.; Hirakawa, H.; Ohshima, K.; Yamashita, A.; Shiba, T.; Ogasawara, N.; Hattori, M.; Kuhara, S.; Hayashi, S. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 2002, 99, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Pridmore, R.D.; Berger, B.; Desiere, F.; Vilanova, D.; Barretto, C.; Pittet, A.C.; Zwallen, M.C.; Rouvet, M.; Altermann, E.; Barrangou, R.; et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 2004, 101, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.J.; Walker, D.H.; Liu, Y.; Zhang, L. Amino acid biosynthesis deficiency in bacteria associated with human and animal hosts. Infect. Genet. Evol. 2009, 9, 514–517. [Google Scholar] [CrossRef]
- Nolling, J.; Breton, G.; Omeichenko, M.V.; Marakova, K.S.; Zeng, Q.; Gibson, R.; Lee, H.M.; Dubois, D.; Qiu, D.; Hitti, J.; et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 2001, 183, 4823–4838. [Google Scholar] [CrossRef]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis spp. Lactis IL1403. Genome. Res. 2011, 11, 731–753. [Google Scholar] [CrossRef]
- Godon, J.J.; Delorme, C.; Bardowski, J.; Chopin, M.C.; Ehrlich, S.D.; Renault, P. Gene inactivation in Lactococcus lactis: Branched-chain amino acid biosynthesis. J. Bacteriol. 1993, 175, 4383–4390. [Google Scholar] [CrossRef]
- Dai, Z.L.; Wu, G.; Zhu, W.Y. Amino acid metabolism in intestinal bacteria: Links between gut ecology and host health. Front. Biosci. 2011, 16, 1768–1786. [Google Scholar] [CrossRef]
- Barker, H.A. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 1981, 50, 23–40. [Google Scholar] [CrossRef]
- Riggottier-Gois, L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef]
- Kim, R.; Attayek, P.J.; Wang, Y.; Furtado, K.L.; Tamayo, R.; Sims, C.E.; Allbritton, N.L. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication 2019, 12, 015006. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tomé, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hetzel, M.; Boiangiu, C.D.; Buckel, W. Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 2004, 28, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Sonnenburg, J.L. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011, 10, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Mei, R.; Nobu, M.K.; Liu, W.T. Identifying anaerobic amino acids degraders through the comparison of short-term and long- term enrichments. Environ. Microbiol. Rep. 2020, 12, 173–184. [Google Scholar] [CrossRef]
- Fonknechten, N.; Chaussonnerie, S.; Tricot, S.; Lajus, A.; Andreesen, J.R.; Perchat, N.; Pelletier, E.; Gouyvenoux, M.; Barbe, V.; Salanoubat, M.; et al. Clostridium stricklandii, a specialist in amino acid degradation: Revisiting its metabolism through its genome sequence. BMC Genom. 2010, 11, 555. [Google Scholar] [CrossRef]
- Birkett, A.; Muir, J.; Phillips, J.; Jones, G.; O’Dea, K. Resistant starch lowers fecal concentrations of ammonia and phenol in humans. Am. J. Clin. Nutr. 1996, 63, 766–772. [Google Scholar] [CrossRef]
- Geboes, K.P.; De Hertogh, G.; De Preter, V.; Luypaerts, A.; Bammens, B.; Evenepoel, P.; Ghoss, Y.; Geboes, K.; Rutgeerts, P.; Verbeke, K. The influence of inulin on the absorption of nitrogen and the production of metabolites of protein fermentation in the colon. Br. J. Nutr. 2006, 96, 1078–1086. [Google Scholar] [CrossRef]
- Windey, K.; De Preter, V.; Huys, G.; Broekaert, W.F.; Delcour, J.A.; Louat, T.; Herman, J.; Verbeke, K. Wheat bran extract alters colonic fermentation and microbial composition but does not affect faecal water toxicity: A randomized controlled trial in healthy subjects. Br. J. Nutr. 2015, 113, 225–238. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T. Role of intestinal bacteria in nutrient metabolism. J. Parenter. Enter. Nutr. 1997, 21, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Pavao, A.; Graham, M.; Arrieta-Ortiz, M.L.; Immanuel, S.R.C.; Baliga, N.S.; Bry, L. Reconsidering the in vivo functions of Clostridial Stickland amino acid fermentations. Anaerobe 2022, 76, 102600. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Gibson, G.R.; Cummings, J.H. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 1992, 72, 57–64. [Google Scholar] [PubMed]
- Zeng, X.; Xing, X.; Gupta, M.; Keber, F.C.; Lopez, J.G.; Lee, Y.J.; Roichman, A.; Wang, L.; Neinast, M.D.; Donia, M.S.; et al. Gut bacterial nutrient preferences quantified in vivo. Cell 2022, 185, 3441–3456.e19. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, S.; Jia, W. Calorie restriction and its impact on gut microbial composition and global metabolism. Front. Med. 2018, 12, 634–644. [Google Scholar] [CrossRef]
- Schmidt, N.S.; Lorentz, A. Dietary restrictions modulate the gut microbiota: Implications for health and disease. Nutr. Res. 2021, 89, 10–22. [Google Scholar] [CrossRef]
- Flint, H.J. The impact of nutrition on the human microbiome. Nutr. Rev. 2012, 70, S10–S13. [Google Scholar] [CrossRef]
- Sbierski-Kind, J.; Grenkowitz, S.; Schlickeiser, S.; Sandforth, A.; Friedrich, M.; Kunkel, D.; Glauben, R.; Brachs, S.; Mai, K.; Thürmer, A.; et al. Effects of caloric restriction on the gut microbiome are linked with immune senescence. Microbiome 2022, 10, 57. [Google Scholar] [CrossRef]
- Kable, M.E.; Chin, E.L.; Storms, D.; Lemay, D.G.; Stephensen, C.B. Tree-based analysis of dietary diversity captures associations between fiber intake and gut microbiota composition in a healthy US adult cohort. J. Nutr. 2022, 152, 779–788. [Google Scholar] [CrossRef]
- Tanes, C.; Bittinger, K.; Gao, Y.; Friedman, E.S.; Nessel, L.; Paladhi, U.R.; Chau, L.; Panfen, E.; Fischbach, M.A.; Braun, J.; et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021, 29, 394–407.e5. [Google Scholar] [CrossRef]
- Beaumont, M.; Portune, K.J.; Steuer, N.; Lan, A.; Cerrudo, V.; Audebert, M.; Dumont, F.; Mancano, G.; Khodorova, N.; Andriamihaja, M.; et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 2017, 106, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Windey, K.; De Preter, V.; Louat, T.; Schuit, F.; Herman, J.; Vansant, G.; Verbeke, K. Modulation of protein fermentation does not affect fecal water toxicity: A randomized cross-over study in healthy subjects. PLoS ONE 2012, 7, e52387. [Google Scholar] [CrossRef] [PubMed]
- Bel Lassen, P.; Belda, E.; Prifti, E.; Dao, M.C.; Specque, F.; Henegar, C.; Rinaldi, L.; Wang, X.; Kennedy, S.P.; Zucker, J.D.; et al. Protein supplementation during an energy-restricted diet induces visceral fat loss and gut microbiota amino acid metabolism activation: A randomized trial. Sci. Rep. 2021, 11, 15620. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, G.V.; Choi, K.; Klemashevich, C.; Wu, C.; Prabakaran, D.; Pan, L.B.; Steimeyer, S.; Mueller, C.; Yousofshani, M.; Alaniz, R.C.; et al. Prediction and quantification of bioactive microbiota metabolites in the mouse. Nat. Commun. 2014, 5, 5492. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Cummings, J.H.; Macfarlane, S.; Gibson, G.R. Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous system. J. Appl. Bacteriol. 1989, 67, 520–527. [Google Scholar] [CrossRef]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gobel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef]
- Liu, X.; Blouin, J.M.; Santacruz, A.; Lan, A.; Andriamihaja, M.; Wilkanowicz, S.; Benetti, P.H.; Tomé, D.; Sanz, Y.; Blachier, F.; et al. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism: The increased luminal bulk connection. Am. J. Physiol. 2014, 307, G459–G470. [Google Scholar] [CrossRef]
- Endo, A.; Nakamura, S.; Konishi, K.; Nakagawa, J.; Tochio, T. Variations in prebiotic oligosaccharide fermentation by intestinal lactic acid bacteria. Int. J. Food Sci. Nutr. 2016, 67, 125–132. [Google Scholar] [CrossRef]
- Weber, F.L., Jr. Effects of lactulose on nitrogen metabolism. Scand. J. Gastroenterol. 1997, 222, 83–87. [Google Scholar] [CrossRef]
- Liong, M.T.; Shah, N.P. Production of organic acids from fermentation of mannitol, fructooligosaccharide and inulin by a cholesterol removing Lactobacillus acidophilus strain. J. Appl. Microbiol. 2005, 99, 783–793. [Google Scholar] [CrossRef]
- Sheridan, P.O.; Louis, P.; Tsompanidou, E.; Shaw, S.; Harmsen, H.J.; Duncan, S.H.; Flint, H.J.; Walker, A.W. Distribution, organization and expression of genes concerned with anaerobic lactate utilization in human intestinal bacteria. Microb. Genom. 2022, 8, 000739. [Google Scholar] [CrossRef] [PubMed]
- Koestler, B.J.; Fisher, C.R.; Payne, S.M. Formate promotes Shigella intercellular spread and virulence gene expression. mBio 2018, 9, e01777-18. [Google Scholar] [CrossRef] [PubMed]
- Shaulov, Y.; Shimokawa, C.; Trebicz-Geffren, M.; Nagaraja, S.; Methling, K.; Lalk, M.; Weiss-Cerem, L.; Lamm, A.T.; Hisaeda, H.; Ankri, S. Escherichia coli mediated resistance of Entamoeba histolytica to oxidative stress is triggered by oxaloacetate. PLoS Pathog. 2018, 14, e1007295. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weiner, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Ariyoshi, T.; Kuroki, Y.; Eguchi, S.; Higashi, S.; Mori, T.; Nonogaki, T.; Iwasaki, K.; Yamashita, M.; Asai, N.; et al. Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci. Rep. 2021, 11, 15007. [Google Scholar] [CrossRef]
- Bone, E.; Tamm, A.; Hill, M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am. J. Clin. Nutr. 1976, 29, 1448–1454. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, fiy125. [Google Scholar] [CrossRef]
- Geypens, B.; Claus, D.; Evenepoel, P.; Hiele, M.; Maes, B.; Peeters, M.; Rutgeers, P.; Ghoos, Y. Influence of dietary protein supplements on the formation of bacterial metabolites in the colon. Gut 1997, 41, 70–76. [Google Scholar] [CrossRef]
- Passmore, I.J.; Letertre, M.P.M.; Preston, M.D.; Bianconi, I.; Harrison, M.A.; Nasher, F.; Kaur, H.; Hong, H.A.; Baines, H.D.; Cutting, S.M.; et al. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS Pathog. 2018, 14, e1007191. [Google Scholar] [CrossRef]
- Harrison, M.A.; Strahl, H.; Dawson, L.F. Regulation of para-cresol production in Clostridioides difficile. Curr. Opin. Microbiol. 2022, 65, 131–137. [Google Scholar] [CrossRef]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Keszthelyi, D.; Troost, F.J.; Masclee, A.A. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol. Motil. 2009, 21, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Wood, T.K.; Lee, J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Rattanaphan, P.; Mittraparp-Arthorn, P.; Srinoun, K.; Vuddhakul, V.; Tansila, N. Indole signaling decreases biofilm formation and related virulence of Listeria monocytogenes. FEMS Microbiol. Lett. 2020, 367, fnaa116. [Google Scholar] [CrossRef]
- Lee, J.; Attila, C.; Cirillo, S.L.; Cirillo, J.D.; Wood, T.K. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2009, 2, 75–90. [Google Scholar] [CrossRef]
- Nikaido, E.; Giraud, E.; Baucheron, S.; Yamasaki, S.; Wiedemann, A.; Okamoto, K.; Takagi, T.; Yamaguchi, A.; Cloeckaert, A.; Nishino, K. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog. 2012, 4, 5. [Google Scholar] [CrossRef]
- Oh, S.; Go, G.W.; Mylonakis, E.; Kim, Y. The bacterial signalling molecule indole attenuates the virulence of the fungal pathogen Candida albicans. J. Appl. Microbiol. 2012, 113, 622–628. [Google Scholar] [CrossRef]
- Nowak, A.; Libudzisz, Z. Influence of phenol, p-cresol and indole on growth and survival of intestinal lactic acid bacteria. Anaerobe 2006, 12, 80–84. [Google Scholar] [CrossRef]
- Ledala, N.; Malik, M.; Rezaul, K.; Paveglio, S.; Provatas, A.; Kiel, A.; Caimano, M.; Zhou, Y.; Lindgren, J.; Krasulova, K.; et al. Bacterial indole as a multifunctional regulator of Klebsella oxytoca complex enterotoxicity. mBio 2022, 13, e0375221. [Google Scholar] [CrossRef]
- Gorelik, O.; Rogad, A.; Holoidovsky, L.; Meijler, M.M.; Sal-Man, N. Indole intercepts the communication between enteropathogenic E. coli and Vibrio cholerae. Gut Microbes 2022, 14, 2138677. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Plants-Paris, K.; Bishoff, D.; DuPont, H.L. Clostridium difficile modulates the gut microbiota by inducing the production of indole, an interkingdom signaling and antimicrobial molecule. mSystems 2019, 4, e00346-18. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, Y.; Oh, S.; Oh, S.; Chun, T.; Kim, T.; Kim, S.H. Inhibitory effect of skatole (3-methylindole) on enterohemorrhagic Escherichia coli O157:H7 ATCC 43894 biofilm formation mediated by elevated endogenous oxidative stress. Lett. Appl. Microbiol. 2014, 58, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Probert, H.M.; Gibson, G.R. Bacterial biofilms in the human gastrointestinal tract. Curr. Issues Intest. Microbiol. 2002, 3, 23–27. [Google Scholar]
- Blachier, F.; Davila, A.M.; Mimoun, S.; Benetti, P.H.; Atanasiu, C.; Andriamihaja, M.; Benamouzig, R.; Bouillaud, F.; Tomé, D. Luminal sulfide and large intestine mucosa: Friend or foe? Amino Acids 2010, 39, 335–347. [Google Scholar] [CrossRef]
- Rowan, F.E.; Docherty, N.G.; Coffey, J.C.; O’Connell, P.R. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br. J. Surg. 2009, 96, 151–158. [Google Scholar] [CrossRef]
- Laue, H.; Denger, K.; Cook, A.M. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl. Environ. Microbiol. 1997, 63, 2016–2021. [Google Scholar] [CrossRef]
- Laue, H.; Friedrich, M.; Ruff, J.; Cook, A.M. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 2001, 183, 1727–1733. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef]
- Yao, C.K.; Rotbart, A.; Ou, J.Z.; Kalantar-Zadeh, K.; Muir, J.G.; Gibson, P.R. Modulation of colonic hydrogen sulfide production by diet and mesalazine utilizing a novel gas-profiling technology. Gut Microbes 2018, 9, 510–522. [Google Scholar] [CrossRef]
- Teigen, L.; Biruete, A.; Khoruts, A. Impact of diet on hydrogen sulfide production: Implications for gut health. Curr. Opin. Clin. Nutr. Metab. Care 2023, 26, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Magee, E.A.; Richardson, C.J.; Hughes, R.; Cummings, J.H. Contribution of dietary protein to sulfide production in the large intestine: An in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 2000, 72, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Bachenheimer, A.G.; Bennett, E.O. The sensitivity of mixed population of bacteria to inhibitors. I. The mechanism by which desulfovibrio desulfuricans protects Ps. aeruginosa from the toxicity of mercurials. Antonie Van Leeuwenhoek 1961, 27, 180–188. [Google Scholar] [CrossRef]
- Stutzenberger, F.J.; Bennett, E.O. Sensitivity of mixed populations of Staphylococcus aureus and Escherichia coli to mercurials. Appl. Microbiol. 1965, 13, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldan, L.; Bellés, A.; Bueno, J.; Azcona-Guttierrez, J.M.; Rojo-Bezares, B.; Torres, C.; Castillo, F.J.; Saenz, Y.; Seral, C. Pseudomonas aeruginosa Isolates from Spanish Children: Occurrence in Faecal Samples, Antimicrobial Resistance, Virulence, and Molecular Typing. Biomed. Res. Int. 2018, 2018, 8060178. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Otto, M. Probiotics to prevent Staphylococcus aureus disease? Gut Microbes 2020, 11, 94–101. [Google Scholar] [CrossRef]
- Pal, V.K.; Bandyopadhyay, P.; Singh, A. Hydrogen sulfide in physiology and pathogenesis of bacteria and viruses. IUBMB Life 2018, 70, 393–410. [Google Scholar] [CrossRef]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A universal defense against antibiotics in bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef]
- Mironov, A.; Seregina, T.; Nagornykh, M.; Luhachack, L.G.; Korolkova, N.; Lopes, L.E.; Kotova, V.; Zavilgelsky, G.; Shakulov, R.; Shatalin, K.; et al. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 6022–6027. [Google Scholar] [CrossRef]
- Shukla, P.; Khodade, V.S.; SharathChandra, M.; Chauhan, P.; Mishra, S.; Siddaramappa, S.; Pradeep, B.E.; Singh, A.; Chakrapani, H. “On demand” redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem. Sci. 2017, 8, 4967–4972. [Google Scholar] [CrossRef]
- Shatalin, K.; Nuthanakanti, A.; Kaushik, A.; Shishov, D.; Peselis, A.; Shamovsky, I.; Pani, B.; Lechpammer, M.; Vasilyev, N.; Shatalina, E.; et al. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science 2021, 372, 1169–1175. [Google Scholar] [CrossRef]
- Ketter, P.M.; Yu, J.J.; Guentzel, M.N.; May, H.C.; Gupta, R.; Eppinger, M.; Klose, K.E.; Seshu, J.; Chambers, J.P.; Cap, A.P.; et al. Acinetobacter baumannii Gastrointestinal Colonization Is Facilitated by Secretory IgA Which Is Reductively Dissociated by Bacterial Thioredoxin A. mBio 2018, 9, e1298-18. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Ong, K.X.; Surendran, S.T.; Sinha, A.; Lai, J.J.H.; Chen, J.; Liang, J.; Tay, L.K.S.; Cui, L.; Loo, H.L.; et al. Hydrogen sulfide sensitizes Acinetobacter baumannii to killing by antibiotics. Front. Microbiol. 2020, 11, 1875. [Google Scholar] [CrossRef] [PubMed]
- Stacy, A.; Andrade-Oliveira, V.; McCulloch, J.A.; Hild, B.; Oh, J.H.; Perez-Chaparro, P.J.; Sim, C.K.; Lim, A.I.; Link, V.M.; Enamorado, M.; et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 2021, 184, 615–627.e17. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Swiatlo, E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010, 42, 39–51. [Google Scholar] [CrossRef]
- Maurelli, A.T.; Fernandez, R.E.; Bloch, C.A.; Rode, C.K.; Fasano, A. “Black holes” and bacterial pathogenicity: A large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 3943–3948. [Google Scholar] [CrossRef]
- Fernandez, I.M.; Silva, M.; Schuch, R.; Walker, W.A.; Siber, A.M.; Maurelli, A.T.; McCormick, B.A. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: A connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 2001, 184, 743–753. [Google Scholar] [CrossRef]
- Burrell, M.; Hanfrey, C.C.; Murray, E.J.; Stanley-Wall, N.R.; Michael, A.J. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J. Biol. Chem. 2010, 285, 39224–39238. [Google Scholar] [CrossRef]
- Sobe, R.C.; Bond, W.G.; Wotanis, C.K.; Zayner, J.P.; Burriss, M.A.; Fernandez, N.; Bruger, E.L.; Waters, C.M.; Neufeld, H.S.; Karatan, E. Spermine inhibits Vibrio cholerae biofilm formation through the NspS-MbaA polyamine signaling system. J. Biol. Chem. 2017, 292, 17025–17036. [Google Scholar] [CrossRef]
- Chagneau, C.V.; Garcie, C.; Bossuet-Greif, N.; Tronnet, S.; Brachmann, A.O.; Piel, J.; Nougayrède, J.P.; Martin, P.; Oswald, E. The polyamine spermidine modulates the production of the bacterial genotoxin colibactin. mSphere 2019, 4, e00414-19. [Google Scholar] [CrossRef] [PubMed]
- Goforth, J.B.; Walter, N.E.; Karatan, E. Effects of polyamines on Vibrio cholerae virulence properties. PLoS ONE 2013, 8, e60765. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Otaru, N.; Ye, K.; Mujezinovic, D.; Berchtold, L.; Constancias, F.; Cornejo, F.A.; Krzystek, A.; de Wouters, T.; Braegger, C.; Lacroix, C.; et al. GABA Production by human intestinal Bacteroides spp.: Prevalence, regulation, and role in acid stress tolerance. Front. Microbiol. 2021, 12, 656895. [Google Scholar] [CrossRef]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Feehily, C.; Karatzas, K.A.G. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef]
- Cotter, P.D.; Hill, C. Surviving the acid test: Responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 2003, 67, 429–453. [Google Scholar] [CrossRef]
- Dicks, L.M.T. Gut Bacteria and Neurotransmitters. Microorganisms 2022, 10, 1838. [Google Scholar] [CrossRef]
- Boyanova, L. Stress hormone epinephrine (adrenaline) and norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe 2017, 44, 13–19. [Google Scholar] [CrossRef]
- Lustri, B.C.; Sperandio, V.; Moreira, C.G. Bacterial chat: Intestinal metabolites and signals in host-microbiota-pathogen interactions. Infect. Immun. 2017, 85, e00476-17. [Google Scholar] [CrossRef]
- O’Donnell, P.M.; Aviles, H.; Lyte, M.; Sonnenfeld, G. Enhancement of in vitro growth of pathogenic bacteria by norepinephrine: Importance of inoculum density and role of transferrin. Appl. Environ. Microbiol. 2006, 72, 5097–5099. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.C.; Vuong, H.E.; Luna, C.D.G.; Pronovost, G.N.; Aleksandrova, A.A.; Riley, N.G.; Vavilina, A.; McGinn, J.; Rendon, T.; Forrest, L.R.; et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 2019, 4, 2064–2073. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, C.; Zhang, G.; Zhan, H.; Liu, B.; Li, C.; Wang, L.; Wang, H.; Wang, J. Antimicrobial mechanism of 4-hydroxyphenylacetic acid on Listeria monocytogenes membrane and virulence. Biochem. Biophys. Res. Commun. 2021, 572, 145–150. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blachier, F. Amino Acid-Derived Bacterial Metabolites in the Colorectal Luminal Fluid: Effects on Microbial Communication, Metabolism, Physiology, and Growth. Microorganisms 2023, 11, 1317. https://doi.org/10.3390/microorganisms11051317
Blachier F. Amino Acid-Derived Bacterial Metabolites in the Colorectal Luminal Fluid: Effects on Microbial Communication, Metabolism, Physiology, and Growth. Microorganisms. 2023; 11(5):1317. https://doi.org/10.3390/microorganisms11051317
Chicago/Turabian StyleBlachier, François. 2023. "Amino Acid-Derived Bacterial Metabolites in the Colorectal Luminal Fluid: Effects on Microbial Communication, Metabolism, Physiology, and Growth" Microorganisms 11, no. 5: 1317. https://doi.org/10.3390/microorganisms11051317
APA StyleBlachier, F. (2023). Amino Acid-Derived Bacterial Metabolites in the Colorectal Luminal Fluid: Effects on Microbial Communication, Metabolism, Physiology, and Growth. Microorganisms, 11(5), 1317. https://doi.org/10.3390/microorganisms11051317