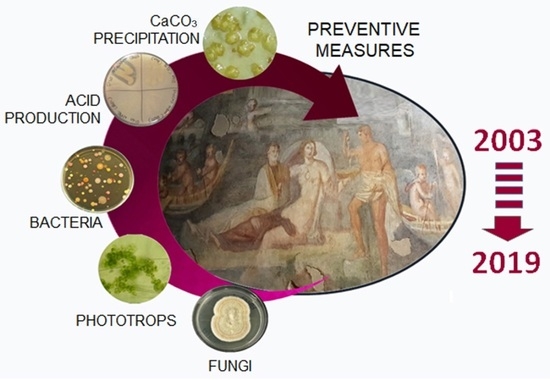
The Roman Houses of the Caelian Hill (Rome, Italy): Multitemporal Evaluation of Biodeterioration Patterns
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site, Previous Intervention, and Analyses
2.2. Present Investigations: Sampling, Environmental Measurements and Biodeteriogens Identification
2.2.1. Field Analysis: Microclimatic Measurements and Sampling
2.2.2. Cultivation, Isolation, and Identification of the BP’ Components
2.3. Multitemporal Analysis
2.4. Plate Assay for Bacterial Detrimental Potential
3. Results
3.1. Current Conservative Conditions
3.2. Identification of Biodeteriogens
3.3. The Multitemporal Evaluation
Total Counts Comparison
3.4. The Assessment of the Detrimental Potential of the Occurring Bacterial Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caneva, G.; Isola, D.; Lee, H.J.; Chung, Y.J. Biological risk for hypogea: Shared data from Etruscan tombs in Italy and ancient tombs of the Baekje dynasty in Republic of Korea. Appl. Sci. 2020, 10, 6104. [Google Scholar] [CrossRef]
- Sanchez-Moral, S.; Luque, L.; Cuezva, S.; Soler, V.; Benavente, D.; Laiz, L.; Gonzalez, J.L.; Sáiz-Jiménez, C. Deterioration of building materials in Roman catacombs: The influence of visitors. Sci. Total Environ. 2005, 349, 260–276. [Google Scholar] [CrossRef]
- Barton, H.A. Introduction to cave microbiology: A review for the non-specialists. J. Cave Karst Stud. 2006, 68, 43–52. [Google Scholar]
- Caneva, G.; Nugari, M.P.; Salvadori, O. Plant Biology for Cultural Heritage: Biodeterioration and Conservation; Getty Publications: Los Angeles, CA, USA, 2008. [Google Scholar]
- Isola, D.; Zucconi, L.; Cecchini, A.; Caneva, G. Dark-pigmented biodeteriogenic fungi in Etruscan hypogeal tombs: New data on their culture-dependent diversity, favouring conditions, and resistance to biocidal treatments. Fungal Biol. 2021, 125, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Caneva, G.; Bartoli, F.; Ceschin, S.; Salvadori, O.; Futagami, Y.; Salvati, L. Exploring ecological relationships in the biodeterioration patterns of Angkor temples (Cambodia) along a forest canopy gradient. J. Cult. Herit. 2015, 16, 728–735. [Google Scholar] [CrossRef]
- Mulec, J. Lampenflora as an accompaniment of mass cave tourism, problems and solutions for Postojnska jama, Slovenia. In The Conservation of Subterranean Cultural Heritage; Saiz-Jimenz, C., Ed.; CRC Press/Balkema: Leiden, The Netherlands; Taylor & Francis Group: London, UK, 2014; pp. 253–256. [Google Scholar]
- Alonso, L.; Pommier, T.; Kaufmann, B.; Dubost, A.; Chapulliot, D.; Doré, J.; Douady, C.J.; Moënne-Loccoz, Y. Anthropization level of Lascaux Cave microbiome shown by regional-scale comparisons of pristine and anthropized caves. Mol. Ecol. 2019, 28, 3383–3394. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Miller, A.Z.; Saiz-Jimenez, C. Lascaux Cave: An example of fragile ecological balance in subterranean environments. In Microbial Life in Cave Systems; Engel, A.S., Ed.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2015; pp. 279–301. [Google Scholar]
- Sun, J.Z.; Ge, Q.Y.; Zhu, Z.B.; Zhang, X.L.; Liu, X.Z. Three dominating hypocrealean fungi of the ‘white mold spots’ on acrylic varnish coatings of the murals in a Koguryo tomb in China. Phytotaxa 2019, 397, 225–236. [Google Scholar] [CrossRef]
- Isola, D.; Bartoli, F.; Langone, S.; Ceschin, S.; Zucconi, L.; Caneva, G. Plant DNA barcode as a tool for root identification in hypogea: The Case of the Etruscan Tombs of Tarquinia (Central Italy). Plants 2021, 10, 1138. [Google Scholar] [CrossRef] [PubMed]
- Zucconi, L.; Canini, F.; Isola, D.; Caneva, G. Fungi affecting wall paintings of historical value: A worldwide meta-analysis of their detected diversity. Appl. Sci. 2022, 12, 2988. [Google Scholar] [CrossRef]
- Bastian, F.; Alabouvette, C. Lights and shadows on the conservation of a rock art cave: The case of Lascaux Cave. Int. J. Speleol. 2009, 38, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Jurado, V.; Fernandez-Cortes, A.; Cuezva, S.; Laiz, L.; Cañaveras, J.C.; Sanchez-Moral, S.; Saiz-Jimenez, C. The fungal colonisation of rock-art caves: Experimental evidence. Naturwissenschaften 2009, 96, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Bruno, L.; Bellezza, S.; De Leo, F.; Urzì, C. A study for monitoring and conservation in the Roman Catacombs of St. Callistus and Domitilla, Rome (Italy). In The Conservation of Subterranean Cultural Heritage; Saiz-Jimenz, C., Ed.; CRC Press/Balkema: Leiden, The Netherlands; Taylor & Francis Group: London, UK, 2014; pp. 37–44. [Google Scholar]
- Sanchez-Moral, S.; Canaveras, J.C.; Laiz, L.; Sáiz-Jiménez, C.; Bedoya, J.; Luque, L. Biomediated precipitation of calcium carbonate metastable phases in hypogean environments: A short review. Geomicrobiol. J. 2003, 20, 491–500. [Google Scholar] [CrossRef] [Green Version]
- Bracci, S.; Cuzman, O.A.; Ignesti, A.; Del Fa, R.M.; Olmi, R.; Pallecchi, P.; Riminesi, C.; Tiano, P. Multidisciplinary approach for the conservation of an Etruscan hypogean monument. Eur. J. Sci. Theol. 2013, 9, 91–106. [Google Scholar]
- Mulec, J. Human impact on underground cultural and natural heritage sites, biological parameters of monitoring and remediation actions for insensitive surfaces: Case of Slovenian show caves. J. Nat. Conserv. 2014, 22, 132–141. [Google Scholar] [CrossRef]
- Bruno, L.; Rugnini, L.; Spizzichino, V.; Caneve, L.; Canini, A.; Ellwood, N.T.W. Biodeterioration of Roman hypogea: The case study of the Catacombs of SS. Marcellino and Pietro (Rome, Italy). Ann. Microbiol. 2019, 69, 1023–1032. [Google Scholar] [CrossRef]
- Soares, F.; Trovão, J.; Portugal, A. Phototrophic and fungal communities inhabiting the Roman cryptoporticus of the national museum Machado de Castro (UNESCO site, Coimbra, Portugal). World J. Microbiol. Biotechnol. 2022, 38, 157. [Google Scholar] [CrossRef]
- Hoyos, M.; Soler, V.; Cañaveras, J.C.; Sánchez-Moral, S.; Sanz-Rubio, E. Microclimatic characterization of a karstic cave: Human impact on microenvironmental parameters of a prehistoric rock art cave (Candamo Cave, northern Spain). Environ. Geol. 1998, 33, 231–242. [Google Scholar] [CrossRef]
- Bartolini, M.; Nugari, M.P.; Pietrini, A.M.; Ricci, S.; Roccardi, A.; Filetici, M.G. Gli ambienti ipogei delle domus romane al Celio. Indagini biologiche per il controllo e la prevenzione del biodeterioramento. Kermes 2010, 77, 45–54. [Google Scholar]
- Bartolini, M.; Nugari, M.P.; Pietrini, A.M.; Ricci, S.; Roccardi, A. Controlli biologici e monitoraggi ambientali per la definizione degli interventi di manutenzione. In Caelius II, Pars Inferior Le Case Romane Sotto la Basilica di Ss. Giovanni e Paolo; Englen, A., Filetici, M.G., Pavolini, C., Santolini, R., Eds.; L’Erma di Bretschneider: Rome, Italy, 2016. [Google Scholar]
- Cecchini, A.; Adamo, F.; Buranelli, F.; Cataldi, M. Le Tombe Dipinte di Tarquinia: Vicenda Conservativa, Restauri, Tecnica di Esecuzione; Nardini Editore: Florence, Italy, 2012. [Google Scholar]
- Albertano, P.; Bruno, L. The importance of light in the conservation of hypogean monuments. In Molecular Biology and Cultural Heritage; Saiz-Jimenez, C., Ed.; Balkema: Lisse, The Netherlands, 2003; pp. 171–177. [Google Scholar]
- Albertano, P.; Bruno, L.; Bellezza, S. New strategies for the monitoring and control of cyanobacterial films on valuable lithic faces. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2005, 139, 311–322. [Google Scholar] [CrossRef]
- Roldán, M.; Oliva, F.; Del Valle, M.G.; Sáiz-Jiménez, C.; Hernández-Mariné, M. Does green light influence the fluorescence properties and structure of phototrophic biofilms? Appl. Environ. Microbiol. 2006, 72, 3026–3031. [Google Scholar] [CrossRef] [Green Version]
- de Luna, J.M.; Molini, D.V.; Fernandez-Balbuena, A.A.; Botella, Á.G.; Herraez, J.A.; Ontañon, R. Selective spectral LED lighting system applied in Paleolithic cave art. Leukos 2015, 11, 223–230. [Google Scholar] [CrossRef]
- Bruno, L.; Valle, V. Effect of white and monochromatic lights on cyanobacteria and biofilms from Roman Catacombs. Int. Biodeterior. Biodegrad. 2017, 123, 286–295. [Google Scholar] [CrossRef]
- Mulec, J. Lampenflora. In Encyclopedia of Caves; Academic Press, Elsevier: Amsterdam, The Netherlands, 2019; pp. 635–641. [Google Scholar]
- Monte, M.; Ferrari, R. Biodeterioration in subterranean environments. Aerobiologia 1993, 9, 141–148. Available online: https://link.springer.com/content/pdf/10.1007/BF02066255.pdf (accessed on 3 March 2023). [CrossRef]
- Agarossi, G.; Ferrari, R.; Monte, M.; Scavizzi, S. Determinazione dell’ecosistema microbico negli ambienti ipogei: Basilica di S. Clemente. In Studi e Ricerche sulla Conservazione delle Opere D’Arte alla Memoria di Marcello Paribeni; Guidobaldi, F., Ed.; CNR: Rome, Italy, 1994; pp. 19–39. [Google Scholar]
- Albertano, P.; Urzì, C. Structural interactions among epilithic cyanobacteria and heterotrophic microorganisms in Roman hypogea. Microb. Ecol. 1999, 38, 244–252. [Google Scholar] [CrossRef]
- Nugari, M.P.; Giuliani, M.P.; Cacace, C. Domus Aurea: Preservation proposal for control of microflora growth of frescoes in hypogean environments. In Biodeterioration of Cultural Property: Proceedings of the International Conference on Biodeterioration of Cultural Property, 20–25 February 1989, held at National Research Laboratory for Conservation of Cultural Property, in Collaboration with ICCROM and INTACH; Macmillan India: Delhi, India, 1991; pp. 359–371. [Google Scholar]
- Albertano, P. Cyanobacterial biofilms in monuments and caves. In Ecology of Cyanobacteria II: Their Diversity in Space and Time; Springer: Dordrecht, The Netherlands, 2012; pp. 317–343. [Google Scholar]
- Urzì, C.; De Leo, F.; Krakova, L.; Pangallo, D.; Bruno, L. Effects of biocide treatments on the biofilm community in Domitilla’s catacombs in Rome. Sci. Total Environ. 2016, 572, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Caneva, G.; Ceschin, S.; De Marco, G. Mapping the risk of damage from tree roots for the conservation of archaeological sites: The case of the Domus Aurea, Rome. Conserv. Manag. Archaeol. Sites 2006, 7, 163–170. [Google Scholar] [CrossRef]
- Caneva, G.; Galotta, G.; Cancellieri, L.; Savo, V. Tree roots and damages in the Jewish catacombs of Villa Torlonia (Roma). J. Cult. Herit. 2009, 10, 53–62. [Google Scholar] [CrossRef]
- Caneva, G.; Langone, S.; Bartoli, F.; Cecchini, A.; Meneghini, C. Vegetation cover and tumuli’s shape as affecting factors of microclimate and biodeterioration risk for the conservation of Etruscan tombs (Tarquinia, Italy). Sustainability 2021, 13, 3393. [Google Scholar] [CrossRef]
- Albertano, P. Epilithic algal communities in hypogean environments. Plant Biosyst. 1993, 127, 386–392. [Google Scholar] [CrossRef]
- Altieri, A.; Pietrini, A.M.; Ricci, S. Un’ associazione di alghe e muschi in un sito archeologico ipogeo. G. Bot. Ital. 1993, 127, 611. [Google Scholar]
- Hernández-Mariné, M.; Clavero, E.; Roldán, M. Why there is such luxurious growth in the hypogean environments? Algol. Stud. /Arch. Für Hydrobiol. 2003, 109, 229–240. [Google Scholar] [CrossRef]
- Bruno, L.; Billi, D.; Urzì, C.; Albertano, P. Genetic characterization of epilithic cyanobacteria and their associated bacteria. Geomicrobiol. J. 2006, 23, 293–299. [Google Scholar] [CrossRef]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C. Epilithic biofilms in Saint Callixtus Catacombs (Rome) harbour a broad spectrum of Acidobacteria. Antonie Van Leeuwenhoek 2006, 89, 203–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Leo, F.; Iero, A.; Zammit, G.; Urzì, C. Chemoorganotrophic bacteria isolated from biodeteriorated surfaces in cave and catacombs. Int. J. Speleol. 2012, 41, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Cuzman, O.A.; Tapete, D.; Fratini, F.; Mazzei, B.; Riminesi, C.; Tiano, P. Assessing and facing the biodeteriogenic presence developed in the Roman Catacombs of Santi Marco, Marcelliano e Damaso, Italy. Eur. J. Sci. Theol. 2014, 10, 185–197. [Google Scholar]
- Krakova, L.; De Leo, F.; Bruno, L.; Pangallo, D.; Urzì, C. Complex bacterial diversity in the white biofilms of the Catacombs of St. Callixtus in Rome evidenced by different investigation strategies. Environ. Microbiol. 2015, 17, 1738–1752. [Google Scholar] [CrossRef]
- Urzi, C.; De Leo, F.; Schumann, P. Kribbella catacumbae sp. nov. and Kribbella sancticallisti sp. nov., isolated from whitish-grey patinas in the catacombs of St Callistus in Rome, Italy. Int. J. Syst. Evol. Microbiol. 2008, 58, 2090–2097. [Google Scholar] [CrossRef] [Green Version]
- Groth, I.; Schumann, P.; Schütze, B.; Gonzalez, J.M.; Laiz, L.; Suihko, M.L.; Stackebrandt, E. Myceligenerans crystallogenes sp. nov., isolated from Roman catacombs. Int. J. Syst. Evol. Microbiol. 2006, 56, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Alias-Villegas, C.; Jurado, V.; Laiz, L.; Miller, A.Z.; Saiz-Jimenez, C. Nocardioides albertanoniae sp. nov., isolated from Roman catacombs. Int. J. Syst. Evol. Microbiol. 2013, 63, 1280–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alias-Villegas, C.; Jurado, V.; Laiz, L.; Saiz-Jimenez, C. Sphingopyxis italica sp. nov., isolated from Roman catacombs. Int. J. Syst. Evol. Microbiol. 2013, 63, 2565–2569. [Google Scholar] [CrossRef] [PubMed]
- Everest, G.J.; Curtis, S.M.; De Leo, F.; Urzì, C.; Meyers, P.R. Kribbella albertanoniae sp. nov., isolated from a Roman catacomb, and emended description of the genus Kribbella. Int. J. Syst. Evol. Microbiol. 2013, 63, 3591–3596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everest, G.J.; Curtis, S.M.; De Leo, F.; Urzì, C.; Meyers, P.R. Description of Kribbella italica sp. nov., isolated from a Roman catacomb. Int. J. Syst. Evol. Microbiol. 2015, 65, 491–496. [Google Scholar] [CrossRef]
- Urzì, C.; De Leo, F.; Bruno, L.; Pangallo, D.; Krakova, L. New species description, biomineralization processes and biocleaning applications of Roman Catacombs-living bacteria. In The Conservation of Subterranean Cultural Heritage; Saiz-Jimenez, C., Ed.; CRC Press/Balkema: Leiden, The Netherlands, 2014; pp. 65–72. [Google Scholar]
- Jurado, V.; Laiz, L.; Sanchez-Moral, S.; Sáiz-Jiménez, C. Pathogenic microorganisms related to human visits in Altamira Cave, Spain. In The Conservation of Subterranean Cultural Heritage; CRC Press/Balkema: Leiden, The Netherlands, 2014; pp. 229–239. [Google Scholar]
- Selbmann, L.; Isola, D.; Egidi, E.; Zucconi, L.; Gueidan, C.; de Hoog, G.S.; Onofri, S. Mountain tips as reservoirs for new rock-fungal entities: Saxomyces gen. nov. and four new species from the Alps. Fungal Divers. 2014, 65, 167–182. [Google Scholar] [CrossRef] [Green Version]
- Isola, D.; Zucconi, L.; Onofri, S.; Caneva, G.; De Hoog, G.S.; Selbmann, L. Extremotolerant rock inhabiting black fungi from Italian monumental sites. Fungal Divers. 2016, 76, 75–96. [Google Scholar] [CrossRef]
- Zammit, G. Systematics and biogeography of sciophilous cyanobacteria; an ecological and molecular description of Albertania skiophila (Leptolyngbyaceae) gen. & sp. nov. Phycologia 2018, 57, 481–491. [Google Scholar]
- Nugari, M.P.; Pietrini, A.M.; Caneva, G.; Imperi, F.; Visca, P. Biodeterioration of mural paintings in a rocky habitat: The Crypt of the Original Sin (Matera, Italy). Int. Biodeterior. Biodegrad. 2009, 63, 705–711. [Google Scholar] [CrossRef]
- Caneva, G.; Bartoli, F.; Fontani, M.; Mazzeschi, D.; Visca, P. Changes in biodeterioration patterns of mural paintings: Multi-temporal mapping for a preventive conservation strategy in the Crypt of the Original Sin (Matera, Italy). J. Cult. Herit. 2019, 40, 59–68. [Google Scholar] [CrossRef]
- Englen, A.; Filetici, M.G.; Palazzo, P.; Pavolini, C.; Astolfi, F.; Alberti, B. Caelius II: Le Case Romane sotto la Basilica dei SS Giovanni e Paolo a Cura di. Collana Palinsesti Romani; L’Erma di Bretschneider: Rome, Italy, 2014; Volume 2. [Google Scholar]
- Normal 9/88 Microflora Autotrofa ed Eterotrofa: Tecniche di Isolamento in Coltura; CNR-ICR: Roma, Italy, 1990.
- Toreno, G.; Isola, D.; Meloni, P.; Carcangiu, G.; Selbmann, L.; Onofri, S.; Caneva, G.; Zucconi, L. Biological colonization on stone monuments: A new low impact cleaning method. J. Cult. Herit. 2018, 30, 100–109. [Google Scholar] [CrossRef]
- Nübel, U.; Ferran, G.P.; Gerard, M. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Valiente Moro, C.; Crouzet, O.; Rasconi, S.; Thouvenot, A.; Coffe, G.; Batisson, I.; Bohatier, J. New design strategy for development of specific primer sets for PCR-based detection of Chlorophyceae and Bacillariophyceae in environmental samples. Appl. Environ. Microbiol. 2009, 75, 5729–5733. [Google Scholar] [CrossRef] [Green Version]
- de Vere, N.; Rich, T.C.; Trinder, S.A.; Long, C. DNA barcoding for plants. In Plant Genotyping; Batley, J., Ed.; Humana Press: New York, NY, USA, 2014; pp. 101–118. [Google Scholar]
- Guiry, M.D.; Guiry, G.M. AlgaeBase Version 4.2. World-Wide Electronic Publication; National University of Ireland: Maynooth, Ireland, 2007. [Google Scholar]
- Wu, F.; Gu, J.D.; Li, J.; Feng, H.; Wang, W. Microbial colonization and protective management of wall paintings. In Cultural Heritage Microbiology: Recent Developments; Archetype Publications: London, UK, 2022. [Google Scholar]
- Sakr, A.A.; Ghaly, M.F.; Edwards, H.G.M.; Ali, M.F.; Abdel-Haliem, M.E. Involvement of Streptomyces in the deterioration of cultural heritage materials through biomineralization and bio-pigment production pathways: A review. Geomicrobiol. J. 2020, 37, 653–662. [Google Scholar] [CrossRef]
- He, D.; Wu, F.; Ma, W.; Gu, J.-D.; Xu, R.; Hu, J.; Yue, Y.; Ma, Q.; Wang, W.; Li, S.-W. Assessment of cleaning techniques and its effectiveness for controlling biodeterioration fungi on wall paintings of Maijishan Grottoes. Int. Biodeterior. Biodegrad 2022, 171, 105406. [Google Scholar] [CrossRef]
- Zammit, G.; De Leo, F.; Urzi, C.; Albertano, P. A non-invasive approach to the polyphasic study of biodeteriogenic biofilms at St Agatha Crypt and Catacombs at Rabat, Malta. In Science and Cultural Heritage in the Mediterranean Area-Diagnostics, Conservation, Experiences and Proposals for a Risk Map; Priulla Srl.: Palermo, Italy, 2009; pp. 323–327. [Google Scholar]
- Baquedano Estevez, C.; Moreno Merino, L.; de la Losa Román, A.; Duran Valsero, J.J. The lampenflora in show caves and its treatment: An emerging ecological problem. Int. J. Speleol. 2019, 48, 4. [Google Scholar] [CrossRef] [Green Version]
- Bartoli, F.; Casanova Municchia, A.; Leotta, M.; Luciano, S.; Caneva, G. Biological recolonization dynamics: Kentridge’s artwork disappearing along the Tiber embankments (Rome, Italy). Int. Biodeterior. Biodegrad. 2021, 160, 105214. [Google Scholar] [CrossRef]
- De Muynck, W.; De Belie, N.; Verstraete, W. Microbial carbonate precipitation in construction materials: A review. Ecol. Eng. 2010, 36, 118–136. [Google Scholar] [CrossRef]
- Jroundi, F.; Fernández-Vivas, A.; Rodriguez-Navarro, C.; Bedmar, E.J.; González-Muñoz, M.T. Bioconservation of deteriorated monumental calcarenite stone and identification of bacteria with carbonatogenic activity. Microb. Ecol. 2010, 60, 39–54. [Google Scholar] [CrossRef]
- Daskalakis, M.I.; Magoulas, A.; Kotoulas, G.; Catsikis, I.; Bakolas, A.; Karageorgis, A.P.; Mavridou, A.; Doulia, D.; Rigas, F. Pseudomonas, Pantoea and Cupriavidus isolates induce calcium carbonate precipitation for biorestoration of ornamental stone. J. Appl. Microbiol. 2013, 115, 409–423. [Google Scholar] [CrossRef]
- Helmi, F.M.; Elmitwalli, H.R.; Elnagdy, S.M.; El-Hagrassy, A.F. Biomineralization consolidation of Fresco wall paintings samples by Bacillus sphaericus. Geomicrobiol. J. 2016, 33, 625–629. [Google Scholar] [CrossRef]
- Zammit, G.; Sánchez-Moral, S.; Albertano, P. Bacterially mediated mineralisation processes lead to biodeterioration of artworks in Maltese catacombs. Sci. Total Environ. 2011, 409, 2773–2782. [Google Scholar] [CrossRef]
- Nigro, L.; Mura, F.; Toti, M.P.; Cirigliano, A.; Rinaldi, T. Carbonatogenic bacteria on the ‘Motya Charioteer’sculpture. J. Cult. Herit. 2022, 57, 256–264. [Google Scholar] [CrossRef]
- Biagioli, F.; Coleine, C.; Piano, E.; Nicolosi, G.; Poli, A.; Prigione, V.; Zanellati, A.; Varese, C.; Isaia, M.; Selbmann, L. Microbial diversity and proxy species for human impact in Italian karst caves. Sci. Rep. 2023, 13, 689. [Google Scholar] [CrossRef]
- Huerta, B.; Marti, E.; Gros, M.; López, P.; Pompêo, M.; Armengol, J.; Barceló, D.; Balcázar, J.L.; Rodríguez-Mozaz, S.; Marcé, R. Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci. Total Environ. 2013, 456, 161–170. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, N.; Shen, X.; Muhammad, A.; Shao, Y. Deciphering environmental resistome and mobilome risks on the stone monument: A reservoir of antimicrobial resistance genes. Sci. Total Environ. 2022, 838, 156443. [Google Scholar] [CrossRef] [PubMed]
- Pinna, D. Coping with Biological Growth on Stone Heritage Objects: Methods, Products, Applications, and Perspectives; Apple Academic Press: Waretown, NJ, USA, 2017. [Google Scholar]
- Ma, W.; Wu, F.; Tian, T.; He, D.; Zhang, Q.; Gu, J.D.; Duan, Y.; Ma, D.; Wang, W.; Feng, H. Fungal diversity and its contribution to the biodeterioration of mural paintings in two 1700-year-old tombs of China. Int. Biodeterior. Biodegrad. 2020, 152, 104972. [Google Scholar] [CrossRef]
- Jurado, V.; Del Rosal, Y.; Liñan, C.; Martin-Pozas, T.; Gonzalez-Pimentel, J.L.; Saiz-Jimenez, C. Diversity and seasonal dynamics of airborne fungi in Nerja Cave, Spain. Appl. Sci. 2021, 11, 6236. [Google Scholar] [CrossRef]
- He, D.; Wu, F.; Ma, W.; Zhang, Y.; Gu, J.D.; Duan, Y.; Xu, R.; Feng, H.; Wang, W.; Li, S.W. Insights into the bacterial and fungal communities and microbiome that causes a microbe outbreak on ancient wall paintings in the Maijishan Grottoes. Int. Biodeterior. Biodegrad. 2021, 163, 105250. [Google Scholar] [CrossRef]
- Velegraki, A.; Cafarchia, C.; Gaitanis, G.; Iatta, R.; Boekhout, T. Malassezia infections in humans and animals: Pathophysiology, detection, and treatment. PLoS Pathog. 2015, 11, e1004523. [Google Scholar] [CrossRef] [Green Version]
- Sert, H.; Sümbül, H.; Sterflinger, K. Microcolonial fungi from antique marbles in Perge/side/Termessos (Antalya/Turkey). Antonie Van Leeuwenhoek 2007, 91, 217–227. [Google Scholar] [CrossRef]
- Dominguez-Moñino, I.; Jurado, V.; Rogerio-Candelera, M.A.; Hermosin, B.; Saiz-Jimenez, C. Airborne fungi in show caves from Southern Spain. Appl. Sci. 2021, 11, 5027. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Medina-Armijo, C.; Isola, D. Black fungi in the built environment—The good, the bad, and the ugly. In Viruses, Bacteria and Fungi in the Built Environment; Woodhead Publishing: Cambridge, UK, 2022; pp. 65–99. [Google Scholar]
- Isola, D.; Bartoli, F.; Caneva, G. The importance of long-term studies after conservative treatments. The case of the Roman Houses of Celius Hill (Rome, Italy). In Proceedings of the 5th European Conference on Biodeterioration of Stone Monuments, Rome, Italy, 11–12 November 2022. [Google Scholar]
- Liu, H.; Li, T.; Ding, Y.; Yang, Y.; Zhao, Z. Dark septate endophytes colonizing the roots of ‘non-mycorrhizal’plants in a mine tailing pond and in a relatively undisturbed environment, Southwest China. J. Plant Interact. 2017, 12, 264–271. [Google Scholar] [CrossRef] [Green Version]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Albano, A.; Alessandrini, A.; Ardenghi, N.M.G.; Astuti, G.; Bacchetta, G.; Ballelli, S.; Banfi, E.; et al. An updated checklist of the vascular flora native to Italy. Plant Biosyst. 2018, 152, 179–303. [Google Scholar] [CrossRef]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments—An updated overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar] [PubMed]
- Ortega-Calvo, J.J.; Ariño, X.; Hernandez-Marine, M.; Saiz-Jimenez, C. Factors affecting the weathering and colonization of monuments by phototrophic microorganisms. Sci. Total Environ. 1995, 167, 329–341. [Google Scholar] [CrossRef]
- Vojtková, H. Algae and their biodegradation effects on building materials in the Ostrava industrial agglomeration. IOP Conf. Ser. Earth Environ. Sci. 2017, 92, 012073. [Google Scholar] [CrossRef]
- Jurado, V.; Gonzalez-Pimentel, J.L.; Fernandez-Cortes, A.; Martin-Pozas, T.; Ontañon, R.; Palacio, E.; Hermosin, B.; Sanchez-Moral, S.; Saiz-Jimenez, C. Early detection of phototrophic biofilms in the polychrome panel, El Castillo Cave, Spain. Appl. Biosci. 2022, 1, 40–63. [Google Scholar] [CrossRef]
- Mulec, J.; Kosi, G. Lampenflora algae and methods of growth control. J. Cave Karst Stud. 2009, 71, 109–115. [Google Scholar]




| Sampling Point | Isolate | Closest BLASTn Match | % Identity | Accession N | CaCO3 | B4 |
|---|---|---|---|---|---|---|
| G1 | B59 | Streptomyces pluricolorescens NBCR12808 | 99.62 | OQ533438 | − | − |
| B82 | Peribacillus simplex NBRC 15720 = DSM 1321 Peribacillus frigoritolerans DSM 8801 Peribacillus castriliensis N3 | 99.10 | OQ606394 | − | ++ | |
| G2 | B103 | Phyllobacterium zundukense Tri-48 Phyllobacterium loti S658 Phyllobacterium trifolii PETP02 | 99.39 | OQ533475 | +++ | + |
| G3 | B58 | Agromyces humi ANK073 | 98.50 | OQ533437 | − | +++ |
| B105 | [Pseudomonas] hibiscola ATCC 19867 Stenotrophomonas maltophilia ATCC 19867 | 99.47 | OQ533476 | − | +++ | |
| B107 | Staphylococcus capitis DSM 20329 | 97.53 | OQ533477 | + | − | |
| B108 | Staphylococcus capitis subsp. capitis DSM 20326 Staphylococcus capitis JCM 2420 | 99.30 | OQ533478 | − | + | |
| G4 | B53 | Cellulomonas cellasea DSM 20118 | 99.62 | OQ533432 | ++ | − |
| G5 | B56 | Alcaligenes faecalis subsp. phenolicus DSM 16503 Alcaligenes javaensis JG3 | 99.59 | OQ533435 | − | +++ |
| B57 | Phyllobacterium endophyticum PEPVI5 | 91.52 | OQ533436 | − | ++ | |
| B61 | Peribacillus castrilensis N3 Peribacillus frigoritolerans DSM 8801 | 97.15 | OQ533440 | − | +++ | |
| B62 | Rhodococcus opacus DSM 4320/ Rhodococcus wratislavensis DSM 44107 | 98.65 | OQ533441 | − | + | |
| B63 | Peribacillus frigotolerans DSMZ 8801 | 100 | OQ533442 | − | ++ | |
| B64 | Psychrobacillus lasiicapitis NEAU-3TGS17 | 98.55 | OQ533443 | − | ++ | |
| B65 | Psychrobacillus lasiicapitis NEAU-3TGS17 | 92.58 | OQ533444 | − | ++ | |
| B66 | Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N3 | 96.69 | OQ533445 | − | ++ | |
| B76a | Microbacterium foliorum DSM 12966 | 98.23 | OQ533454 | − | ++ | |
| B76b | Micrococcus luteus NCTC 2665 Micrococcus yunnanensis YIM 65004 | 95.11 | OQ606393 | − | ++ | |
| Gx | B80 | Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N3 | 98.42 | OQ533458 | − | + |
| P6 | B79 | Amycolatopsis jiguanensis CFHS01580 | 98.96 | OQ533457 | − | ++ |
| B83 | Paenibacillus mobilis S8/ Paenibacillus tundrae A10b | 98.81 | OQ533460 | ++ | + | |
| B112 | Agromyces terreus DS-10 | 99.11 | OQ533479 | − | − | |
| P7 | B37 | Lysinibacillus fusiformis NBRC 15717 | 99.04 | OQ512987 | − | ++ |
| B44 | Lysinibacillus fusiformis NBRC 15717 | 99.00 | OQ533428 | − | +++ | |
| B89 | Promicromonospora xylanilytica YIM61515 | 99.27 | OQ533466 | − | ++ | |
| B97 | Micrococcus endophyticus YIM 56238 | 97.37 | OQ533472 | − | + | |
| P8 | B60 | Advenella kashmirensis WT001 | 98.43 | OQ533439 | ++ | +/− |
| B67 | Phyllobacterium zundukense Tri-48 Phyllobacterium sophorae CCBAU 03422 Phyllobacterium loti S658 | 96.81 | OQ533446 | − | + | |
| B71 | Advenella kashmirensis WT001 | 95.01 | OQ533449 | ++ | ++ | |
| B81 | Peribacillus simplex NBRC 15720 = DSM 1321 Peribacillus frigoritolerans DSM 8801 | 98.78 | OQ533459 | − | ++ | |
| B85 | Phyllobacterium zundukense Tri-48 Phyllobacterium loti S658 Phyllobacterium trifolii PETP02 | 98.64 | OQ533462 | ++ | + | |
| B92 | Microlunatus parietis CCM 7636 | 99.27 | OQ533468 | − | − | |
| B102 | Microlunatus nigridraconis CPCC 203993/ Microlunatus parietis CCM 7636 | 99.31 | OQ533474 | − | + | |
| P9 | B13 | Mesorhizobium atlanticum CNPSo 3140 Mesorhizobium comanense 3P27G6 Mesorhizobium plurifarium NBRC 102498 | 96.81 | OQ512977 | − | |
| B15 | Delftia acidovorans JCM 5833 | 99.45 | OQ512978 | − | + | |
| P10 | B04 | Achromobacter pestifer LMG 3431 Achromobacter piechaudii CCUG 724 | 96.39 | OQ512006 | − | ++ |
| B06 | Achromobacter xylosoxidans FDAARGOS_789 | 99.37 | OQ512007 | − | +++ | |
| B07 | Bacillus thuringiensis ATCC 10792 | 99.25 | OQ512008 | + | − | |
| B08 | Brevundimonas diminuta b71 | 98.52 | OQ512009 | − | ++ | |
| B09 | Nocardia ninae DSM 44978 | 94.95 | OQ512010 | − | +++ | |
| B10 | Achromobacter insolitus NCTC13520 Achromobacter spanius DSM 23806 Achromobacter deleyi LMG 3458 | 99.40 | OQ512011 | − | + | |
| B11 | Delftia acidovorans JCM 5833 | 99.09 | OQ512975 | − | ++ | |
| B12 b | Stenotrophomonas geniculata ATCC 19374 = JCM 13324 | 99.40 | OQ512976 | − | ++ | |
| B41 | Bacillus altitudinis 41KF2b Bacillus aerophilus 28K Bacillus australimaris MCCC 1A05787 Bacillus aerius 24K | 98.78 | OQ533427 | + | + | |
| B54 | Peribacillus castrilensis N3 Peribacillus frigoritolerans DSM 8801 | 99.23 | OQ533433 | − | ++ | |
| B55 | Phyllobacterium zundukense Tri-48 Phyllobacterium trifolii PETP02 | 99.45 | OQ533434 | + | ++ | |
| B70 | Lysinibacillus fusiformis NBRC 15717 | 99.05 | OQ533448 | − | +++ | |
| B74 | Peribacillus frigoritolerans DSM 8801 | 98.13 | OQ533452 | − | ++ | |
| B84 | Phyllobacterium zundukense TRi-48 Phyllobacterium loti S658 Phyllobacterium trifolii PETP02 Phyllobacterium bourgognense STM 201 | 99.13 | OQ533461 | ++ | ++ | |
| B86 | Phyllobacterium endophyticum PEPV15 Phyllobacterium zundukense Tri-48 Phyllobacterium loti S658 | 98.05 | OQ533463 | ++ | ++ | |
| PX | B50 | Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N3 | 99.17 | OQ533430 | − | + |
| B72 | Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N4 | 99.43 | OQ533450 | − | ++ | |
| B75 | Peribacillus simplex NBRC 15720 = DSM 1321 Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N5 | 99.27 | OQ533453 | − | ++ | |
| B77 | Peribacillus castrilensis N3/ Peribacillus frigoritolerans DSM 8801 | 99.63 | OQ533455 | − | + | |
| B78 | Inquilinus ginsengisoli Gsoil 080 | 97.81 | OQ533456 | + | ++ | |
| P11 | B25 | Myroides odoratus FDAARGOS_1131 | 96.76 | OQ512979 | + | − |
| B26 | Providencia rettgeri FDAARGOS 1450 | 98.20 | OQ512980 | + | + | |
| B27 | Providencia rettgeri FDAARGOS 1450 | 98.45 | OQ512982 | + | +/− | |
| B28 | Providencia rettgeri FDAARGOS 1450 | 98.42 | OQ512983 | + | + | |
| B30 | Myroides odoratus DSM 2801 NBRC 14945 | 98.51 | OQ512984 | + | − | |
| B35 | Lysinibacillus fusiformis NBRC 15717 | 98.63 | OQ512985 | − | ++ | |
| B36 | Lysinibacillus fusiformis NBRC 15717 | 99.87 | OQ512986 | − | ++ | |
| B38 | Lysinibacillus fusiformis NBRC 15717 | 99.91 | OQ607474 | − | ++ | |
| B20, B40 | Peribacillus castrilensis N3 Peribacillus frigoritolerans DSM 8801 | 99.37 | OQ512988 | − | ++ | |
| B87 | Inquilinus ginsengisoli Gsoil 080 | 98.91 | OQ533464 | ++ | − | |
| B88 | Promicromonospora kermanensis UTMC 533 | 99.29 | OQ533465 | − | ++ | |
| B90 | Inquilinus ginsegisoli GSOIL 080 | 98.98 | OQ533467 | − | + | |
| B94 | Microbacterium shaanxiense CCNWSP60 Microbacterium arthrosphaerae CCM 7681 Microbacterium murale 01-Gi-001 | 99.33 | OQ533470 | − | ++ | |
| B95 | Leucobacter aerolatus CCM 7705 | 97.37 | OQ533471 | − | +++ | |
| B96 | Serratia liquefaciens ATCC 27592 | 98.61 | OQ606395 | − | − | |
| B125 | Achromobacter insolitus NCTC13520 Achromobacter spanius DSM 23806 | 99.48 | OQ533487 | − | +++ | |
| W-11/b | B116 | Microlunatus parietis 12-Be-011 | 98.66 | OQ533482 | + | + |
| B123 | Paenibacillus lautus NBRC 15380 | 96.69 | OQ533486 | +++ | − | |
| W-A | B150 | Micromonospora cremea CR30 | 98.93 | OQ533480 | + | + |
| B151 | Micromonospora coriariae DSM 44875 Micromonospora cremea CR30 | 98.81 | OQ533481 | + | + | |
| W-B | B121 | Nocardioides panzhihuensis KLBMP 1050 | 98.28 | OQ533485 | − | ++ |
| B153 | Leucobacter salsicius M1-8 Leucobacter exalbidus K-540B | 99.14 | OQ533489 | ++ | − | |
| BALN | B48 | Peribacillus castrilensis N3 Peribacillus frigoritolerans DSM 8801 | 98.53 | OQ533429 | − | ++ |
| B52 | Peribacillus frigoritolerans DSM 8801 Peribacillus castrilensis N4 | 99.72 | OQ533431 | − | +++ | |
| B69 | Lysinibacillus cavernae SYSU K30005/ Lysinibacillus fusiformis NBRC 15717/ Lysinibacillus pakistanensis NCCP-54 | 97.18 | OQ533447 | ++ | − | |
| B73 | Acinetobacter iwoffii FDAARGOS 1393 | 99.47 | OQ533451 | − | + | |
| B93 | Prolinoborus fasciculus CIP 103579 | 98.30 | OQ533469 | − | +/− | |
| B98 | Advenella mimigardefordensis DPN7/ Advenella kashmirensis subsp. methylica PK1 | 97.99 | OQ533473 | − | ++ | |
| B119 | Metabacillus sediminilitoris DSL-17 | 99.52 | OQ533483 | − | ++ | |
| B120 | Nocardioides panzhihuaensis KLBMP1050 | 98.72 | OQ533484 | +/− | + | |
| B-14 | B128 | Leucobacter aerolatus CCM 7705/ Leucobacter salsicius M1-8 | 98.74 | OQ533488 | ++ | − |
| Sampling Point | Isolate | Closest BLASTn Match | % Identity | Accession N |
|---|---|---|---|---|
| G1 | F13 | Penicillium polonicum KMM4719 | 99.02 | OQ512946 |
| G2 | F29 | Aspergillus pseudoglaucus CBS 126221 Aspergillus ruber CBS 126220 Aspergillus glaucus CBS 126.55 | 99.81 99.81 99.81 | OQ512947 |
| F06 | Penicillium oxalicum DUCC5744 | 97.82 | OQ512948 | |
| G3 | F19 | Cladosporium neolangeronii CPC 22267 Cladosporium psychrotolerans DTO:305-G3 Cladosporium langeronii CPC 22326 | 100 100 100 | OQ512949 |
| G4 | F23 | Penicillium chrysogenum NBPen2012A02 | 99.33 | OQ512950 |
| F14 | Chrysosporium undulatum CBS 964.97 | 99.84 | OQ512951 | |
| F22 | Torula hollandica CBS 220.69 | 99.18 | OQ512952 | |
| G5 | F28 | Pseudogymnoascus pannorum CBS 126913 | 91.60 | OQ512953 |
| GX | F38 | Cladosporium halotolerans CBS 127370 Cladosporium parahalotolerans CPC 22373 | 99.43 99.43 | OQ512954 |
| F39 | Penicillium chrysogenum SCSGAF0070 | 99.34 | OQ512955 | |
| F31 | [Coniosporium] MA 4640 | 97.18 | OQ512956 | |
| P6 | F08 | Penicillium brevicompactum CBS 287.53 | 100 | OQ512957 |
| P7 | F32 | Penicillium dipodomyus CBS 110412 Penicillium flavigenum CBS 419.89 Penicillium lanosocoeruleum CBS 334.48 | 97.90 97.90 97.90 | OQ512958 |
| P8 | F17 | Cladosporium halotolerans CBS 114065 | 99.84 | OQ512959 |
| F04 | Cladosporium parahalotolerans CPC 22373 Cladosporium halotolerans CBS 127370 | 99.81 99.81 | OQ512960 | |
| P9 | F01 | Malassezia restricta CBS 7991 | 98.42 | OQ512961 |
| F02 | Penicillium brevicompactum CBS 287.53 Penicillium kongii AS3.15329 | 100 100 | OQ512962 | |
| F09a | Cyphellophora olivacea CCFEE 9916 | 99.33 | OQ512963 | |
| P10 | F18 | Cladosporium endophyticum MFLUCC 17-0599 | 98.46 | OQ512964 |
| P11 | F05 | Cladosporium herbarum CBS 128234 Cladosporium macrocarpum CBS 12778 Cladosporium variabile CBS 121635 Cladosporium ossifragi CBS 842.91 Cladosporium allicinum CBS 188.54 | 99.61 99.61 99.61 99.61 99.61 | OQ512965 |
| Px | F11 | Penicillium rubens CBS 132206 Penicillium chrysogenum CBS 127368 | 99.82 99.82 | OQ512966 |
| W11 | F34 | Penicillium rubens CBS 132206 Penicillium chrysogenum NEF9 | 97.03 97.03 | OQ512967 |
| CV- A | F36 | Penicillium chrysogenum CBS 127368 Penicillium rubens CBS 205.57 | 99.09 99.09 | OQ512968 |
| CV- B | F37 | Penicillium rubens CBS 132206 Penicillium oxalicum DUCC 5744 | 97.44 97.44 | OQ512969 |
| BAL14 | F40 | Cladosporium subuliforme CBS 126500 C. verrucocladosporioides CBS 126363 Cladosporium xylophilum CBS 125997 | 99.81 99.81 99.81 | OQ512970 |
| BAL15 | F41 | Penicillium goetzi CBS 285.73 Penicillium rubens CBS 129667 | 99.07 99.07 | OQ512971 |
| Room | Site | 2002 | 2003 | July 2019 | Nov 2019 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Photot | Fungi | Bacteria | Photot | Fungi | Bacteria | Photot. | Fungi | Bacteria | Photot. | Fungi | Bacteria | ||
| CG | G1 | − | / | / | − | / | / | − | +++ | − | − | +++ | − |
| G2 | − | / | / | − | / | / | − | +++ | − | − | +++ | − | |
| G3 | / | + | + | / | ++ | + | / | − | ++ | / | ++ | − | |
| G4 | / | +++ | − | / | ++ | ++ | / | ++ | − | / | + | +++ | |
| G5 | − | +++ | ++++ | − | − | +++ | − | + | ++ | − | + | +++ | |
| NP | P6 | / | − | − | / | + | ++ | / | − | ++ | / | + | + |
| P7 | − | − | + | − | + | + | − | ++ | + | − | − | ++ | |
| P8 | / | − | − | / | + | − | / | ++ | ++ | / | ++ | − | |
| P9 | / | + | − | / | + | + | / | +++ | + | / | ++ | − | |
| P10 | ++ | + | ++++ | − | − | +++ | − | − | ++ | − | +/− | ++ | |
| W | 11 | / | + | + | / | + | + | / | − | + | / | − | + |
| 11/b | / | + | + | / | + | + | / | − | + | / | − | + | |
| A | − | / | / | − | / | / | +++ | + | + | +++ | + | + | |
| B | − | / | / | − | / | / | +++ | + | + | +++ | + | + | |
| BAL | 14 | / | +++ | ++ | / | +++ | +++ | + | + | ++ | + | + | ++ |
| 15 | / | +++ | ++ | / | +++ | +++ | + | + | ++ | + | + | ++ | |
| G | +++ | / | / | +++ | / | / | + | − | + | + | − | + | |
| Room | Sample | Deter. Pattern | 2002 | 2003 | Deter. Pattern | 2019 | |||
|---|---|---|---|---|---|---|---|---|---|
| NP | P10 | GBP | Navicula sp. | + | − | − | − | ||
| Px | / | / | LGP | Scytonema sp. | +++ | ||||
| W | A | GP | − | − | GP | (*) Pseudostichococcus cfr. Monallardoides Scytonema sp. | +++ +/− | ||
| B | YP | − | − | GP | Chlorella vulgaris | +++ | |||
| BAL | 14 | / | / | FGP | (*) Albertania cfr. skiophila (*) Coccomyxa sp. | ++ + | |||
| 15 | / | / | FGP | (*) Albertania sp. Phormidium laminosum | ++ +++ | ||||
| G | DGP | Plectonema gracillium Desmococcus sp. Navicula gallica Moss protonemata | +++ ++ + ++ | Plectonema gracillium Desmococcus sp. Navicula gallica Chlorococcum sp. Stichococcus bacillaris Moss protonemata | +++ ++ + + + ++ | − | Desmococcus vulgaris Fern’ spores | +++ ++ | |
| By | / | / | GOP | Moss gametophyte | +++ | ||||
| Bx | / | / | DGP | Chlorococcum vulgaris Stichococcus sp. | +++ + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isola, D.; Bartoli, F.; Morretta, S.; Caneva, G. The Roman Houses of the Caelian Hill (Rome, Italy): Multitemporal Evaluation of Biodeterioration Patterns. Microorganisms 2023, 11, 1770. https://doi.org/10.3390/microorganisms11071770
Isola D, Bartoli F, Morretta S, Caneva G. The Roman Houses of the Caelian Hill (Rome, Italy): Multitemporal Evaluation of Biodeterioration Patterns. Microorganisms. 2023; 11(7):1770. https://doi.org/10.3390/microorganisms11071770
Chicago/Turabian StyleIsola, Daniela, Flavia Bartoli, Simona Morretta, and Giulia Caneva. 2023. "The Roman Houses of the Caelian Hill (Rome, Italy): Multitemporal Evaluation of Biodeterioration Patterns" Microorganisms 11, no. 7: 1770. https://doi.org/10.3390/microorganisms11071770
APA StyleIsola, D., Bartoli, F., Morretta, S., & Caneva, G. (2023). The Roman Houses of the Caelian Hill (Rome, Italy): Multitemporal Evaluation of Biodeterioration Patterns. Microorganisms, 11(7), 1770. https://doi.org/10.3390/microorganisms11071770