Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41
Abstract
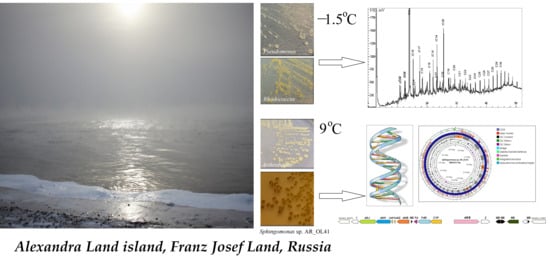
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Media Composition
2.3. A Model Experiment for Cleaning Sand from Oil
2.4. Analytical Methods
2.5. DNA Extraction and 16S rRNA Gene and Genome Sequencing
2.6. Bioinformatic Methods
2.7. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Characterization of the Strains
3.2. Crude Oil Degradation
3.3. Genome Statistics and Functional Characterization of the Sphingomonas sp. AR_OL41
3.4. alkB Genes in Genomes of Sphingomonadaceae Bacteria
3.5. Cold Stress Protection Genes in the Genome of Sphingomonas sp. AR_OL41
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henderson, J.; Loe, J. The prospects and challenges for arctic oil development. Oxf. Inst. Energy Stud. 2014, 54, 1–60. [Google Scholar]
- Bashkin, V.N.; Galiulin, R.V. Innovative geoecological risk assessment in technogenesis for green economy progress. Green Econ. Era Fourth Ind. Revolut. 2021, 1, 45–67. [Google Scholar] [CrossRef]
- Mohn, W.W.; Stewart, G.R. Limiting factors for hydrocarbon biodegradation at low temperature in Arctic soils. Soil Biol. Biochem. 2000, 32, 1161–1172. [Google Scholar] [CrossRef]
- Naseri, M.; Barabadi, A.; Barabady, J. Bioremediation treatment of hydrocarbon-contaminated Arctic soils: Influencing parameters. Environ. Sci. Pollut. Res. Int. 2014, 21, 11250–11265. [Google Scholar] [CrossRef] [PubMed]
- Vasilyeva, G.; Kondrashina, V.; Strijakova, E.; Ortega-Calvo, J.J. Adsorptive bioremediation of soil highly contaminated with crude oil. Sci. Total Environ. 2020, 706, 135739. [Google Scholar] [CrossRef] [PubMed]
- Ławniczak, Ł.; Wózniak-Karczewska, M.; Loibner, A.P.; Heipieper, H.J.; Chrzanowski, Ł. Microbial degradation of hydrocarbons—Basic principles for bioremediation: A Review. Molecules 2020, 25, 856. [Google Scholar] [CrossRef] [PubMed]
- Van Stempvoort, D.; Biggar, K. Potential for bioremediation of petroleum hydrocarbons in groundwater under cold climate conditions: A review. Cold Reg. Sci. Technol. 2008, 53, 16–41. [Google Scholar] [CrossRef]
- Greer, C.W.; Whyte, L.G.; Niederberger, T.D. Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 2313–2328. [Google Scholar] [CrossRef]
- Bell, T.H.; Yergeau, E.; Maynard, C.; Juck, D.; Whyte, L.G.; Greer, C.W. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J. 2013, 7, 1200–1210. [Google Scholar] [CrossRef]
- Mohammed, D.; Hicham, E.A.; Naima, E.G. Biodegradation of environmental pollutants by marine yeasts. In Marine Organisms: A Solution to Environmental Pollution? Environmental Challenges and Solutions; Encarnação, T., Canelas Pais, A., Eds.; Springer: Cham, Switzerland, 2023; pp. 79–91. [Google Scholar] [CrossRef]
- Myazin, V.A.; Korneykova, M.V.; Chaporgina, A.A.; Fokina, N.V.; Vasilyeva, G.K. The Effectiveness of biostimulation, bioaugmentation and sorption-biological treatment of soil contaminated with petroleum products in the Russian Subarctic. Microorganisms 2021, 9, 1722. [Google Scholar] [CrossRef]
- Namsaraev, Z.; Bobrik, A.; Kozlova, A.; Krylova, A.; Rudenko, A.; Mitina, A.; Saburov, A.; Patrushev, M.; Karnachuk, O.; Toshchakov, S. Carbon emission and biodiversity of Arctic soil microbial communities of the Novaya Zemlya and Franz Josef Land Archipelagos. Microorganisms 2023, 11, 482. [Google Scholar] [CrossRef]
- Semenova, E.M.; Babich, T.L.; Sokolova, D.S.; Ershov, A.P.; Raievska, Y.I.; Bidzhieva, S.K.; Stepanov, A.L.; Korneykova, M.V.; Myazin, V.A.; Nazina, T.N. Microbial communities of seawater and coastal soil of Russian Arctic region and their potential for bioremediation from hydrocarbon pollutants. Microorganisms 2022, 10, 1490. [Google Scholar] [CrossRef]
- Korneykova, M.V.; Myazin, V.A.; Fokina, N.V.; Chaporgina, A.A.; Nikitin, D.A.; Dolgikh, A.V. Structure of microbial communities and biological activity in tundra soils of the Euro-Arctic region (Rybachy peninsula, Russia). Microorganisms 2023, 11, 1352. [Google Scholar] [CrossRef]
- Rike, A.G.; Haugen, K.B.; Børresen, M.; Engene, B.; Kolstad, P. In situ biodegradation of petroleum hydrocarbons in frozen arctic soils. Cold Reg. Sci. Technol. 2003, 37, 97–120. [Google Scholar] [CrossRef]
- Rike, A.G.; Haugen, K.B.; Engene, B. In situ biodegradation of hydrocarbons in arctic soil at sub-zero temperatures–field monitoring and theoretical simulation of the microbial activation temperature at a Spitsbergen contaminated site. Cold Reg. Sci. Technol. 2005, 41, 189–209. [Google Scholar] [CrossRef]
- Rivkina, E.M.; Friedmann, E.I.; McKay, C.P.; Gilichinsky, D.A. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 2000, 66, 3230–3233. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Bonaunet, K. Biodegradation of petroleum hydrocarbons in seawater at low temperatures (0–5 °C) and bacterial communities associated with degradation. Biodegradation 2006, 17, 71–82. [Google Scholar] [CrossRef]
- McFarlin, K.M.; Prince, R.C.; Perkins, R.; Leigh, M.B. Biodegradation of dispersed oil in Arctic seawater at −1°C. PLoS ONE 2014, 9, e84297. [Google Scholar] [CrossRef]
- Jesus, H.E.; Carreira, R.S.; Paiva, S.S.M.; Massone, C.; Enrich-Prast, A.; Peixoto, R.S.; Rodrigues, J.L.M.; Lee, C.K.; Cary, C.; Rosado, A.S. Microbial succession under freeze–thaw events and its potential for hydrocarbon degradation in nutrient-amended Antarctic soil. Microorganisms 2021, 9, 609. [Google Scholar] [CrossRef]
- Gerdes, B.; Brinkmeyer, R.; Dieckmann, G.; Helmke, E. Influence of crude oil on changes of bacterial communities in Arctic sea-ice. FEMS Microbiol. Ecol. 2005, 53, 129–139. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y.; Canaguier, R.; Svecova, E.; Cardarelli, M. Biostimulant action of a plant-derived protein hydrolysate produced through enzymatic hydrolysis. Front. Plant Sci. 2014, 5, 448. [Google Scholar] [CrossRef] [PubMed]
- Colla, T.S.; Andreazza, R.; Bücker, F.; de Souza, M.M.; Tramontini, L.; Prado, G.R.; Frazzon, A.P.G.; Camargo, F.A.; Bento, F.M. Bioremediation assessment of diesel–biodiesel-contaminated soil using an alternative bioaugmentation strategy. Environ. Sci. Pollut. Res. 2014, 21, 2592–2602. [Google Scholar] [CrossRef]
- Myazin, V.A.; Isakova, E.A.; Vasilyeva, G.K. Influence of granular activated carbon on the rate of bioremediation of soils in the Murmansk region, historically contaminated with oil products. Probl. Reg. Ekol. 2020, 2, 20–26. [Google Scholar] [CrossRef]
- Myazin, V.A.; Ivanova, L.A.; Chaporgina, A.A.; Fokina, N.V.; Korneikova, M.V.; Evdokimova, G.A. It’s Time to Heal the Arctic. Biological Methods of Cleaning and Remediation of Oil-Contaminated Areas. FRS KSC RAS, Apatity. 2023. Available online: https://rio.ksc.ru/data/documents/2_2022_13/%D0%BF%D0%BE%D1%80%D0%B0%20%D0%BE%D0%B7%D0%B4%D0%BE%D1%80%D0%B0%D0%B2%D0%BB%D0%B8%D0%B2%D0%B0%D1%82%D1%8C%20%D0%B0%D1%80%D0%BA%D1%82%D0%B8%D0%BA%D1%83_2023-%D0%905.pdf (accessed on 1 August 2023).
- Evdokimova, G.A.; Gershenkop, A.S.; Fokina, N.V. The impact of bacteria of circulating water on apatite-nepheline ore flotation. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2012, 47, 398–404. [Google Scholar] [CrossRef]
- Semenova, E.M.; Babich, T.L.; Sokolova, D.S.; Dobriansky, A.S.; Korzun, A.V.; Kryukov, D.R. Microbial diversity of hydrocarbon-contaminated soils of the Franz Josef land Archipelago. Microbiology 2021, 90, 721–730. [Google Scholar] [CrossRef]
- Newsham, K.K.; Danielsen, B.K.; Biersma, E.M.; Elberling, B.; Hillyard, G.; Kumari, P.; Priemé, A.; Woo, C.; Yamamoto, N. Rapid response to experimental warming of a microbial community inhabiting high Arctic patterned ground soil. Biology 2022, 11, 1819. [Google Scholar] [CrossRef]
- Peeb, A.; Dang, N.P.; Truu, M.; Nõlvak, H.; Petrich, C.; Truu, J. Assessment of hydrocarbon degradation potential in microbial communities in Arctic Sea ice. Microorganisms 2022, 10, 328. [Google Scholar] [CrossRef]
- Whyte, L.G.; Bourbonnie`re, L.; Greer, C.W. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 1997, 63, 3719–3723. [Google Scholar] [CrossRef]
- Whyte, L.G.; Slagman, S.J.; Pietrantonio, F.; Bourbonnière, L.; Koval, S.F.; Lawrence, J.R.; Inniss, W.E.; Greer, C.W. Physiological adaptations involved in alkane assimilation at a low temperature by Rhodococcus sp. strain Q15. Appl. Environ. Microbiol. 1999, 65, 2961–2969. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, B.; Phale, P.S. Microbial degradation of naphthalene and substituted naphthalenes: Metabolic diversity and genomic insight for bioremediation. Front. Bioeng. Biotechnol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Pandolfo, E.; Caracciolo, A.B.; Rolando, L. Recent advances in bacterial degradation of hydrocarbons. Water 2023, 15, 375. [Google Scholar] [CrossRef]
- Margesin, R.; Zhang, D.; Busse, H. Sphingomonas alpina sp. nov., a psychrophilic bacterium isolated from alpine soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1558–1563. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Zhang, Y.; Han, S.; Xu, H. Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Environ. Microbiol. 2016, 47, 271–278. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Somee, M.R.; Amoozegar, M.A.; Dastgheib, S.M.M.; Shavandi, M.; Maman, L.G.; Bertilsson, S.; Mehrshad, M. Genome-resolved analyses show an extensive diversification in key aerobic hydrocarbon-degrading enzymes across bacteria and archaea. BMC Genom. 2022, 6, 690. [Google Scholar] [CrossRef]
- Pfennig, N.; Lippert, K.D. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch. Mikrobiol. 1966, 55, 245–256. [Google Scholar] [CrossRef]
- Sokolova, D.S.; Semenova, E.M.; Grouzdev, D.S.; Bidzhieva, S.K.; Babich, T.L.; Loiko, N.G.; Ershov, A.P.; Kadnikov, V.V.; Beletsky, A.V.; Mardanov, A.V.; et al. Sulfidogenic microbial communities of the Uzen high-temperature oil field in Kazakhstan. Microorganisms 2021, 9, 1818. [Google Scholar] [CrossRef]
- Appelbaum, P.C.; Stavitz, J.; Bentz, M.S.; von Kuster, L.C. Comparison of four methods for identification of Gram-negative non-fermenters: Organisms less commonly encountered in clinical specimens. J. Clin. Microbiol. 1980, 12, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.M.; Ershov, A.P.; Sokolova, D.S.; Tourova, T.P.; Nazina, T.N. Diversity and biotechnological potential of nitrate-reducing bacteria from heavy-oil reservoirs (Russia). Microbiology 2020, 89, 685–696. [Google Scholar] [CrossRef]
- Manucharova, N.A.; Bolshakova, M.A.; Babich, T.L.; Tourova, T.P.; Semenova, E.M.; Yanovich, A.S.; Poltaraus, A.B.; Stepanov, A.L.; Nazina, T.N. Microbial degraders of petroleum and polycyclic aromatic hydrocarbons from sod-podzolic soil. Microbiology 2021, 90, 743–753. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Wang, F.; Wei, D.; Wang, X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J. Microbiol. Meth. 2010, 82, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Plakunov, V.K.; Mart’yanov, S.V.; Teteneva, N.A.; Zhurina, M.V. A universal method for quantitative characterization of growth and metabolic activity of microbial biofilms in static models. Microbiology 2016, 85, 509–513. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Windowed Adaptive Trimming for Fastq Files Using Quality. Available online: https://github.com/najoshi/sickle (accessed on 28 January 2023).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Tatusova, T.; Di Cuccio, M.; Badretdin, A.; Chetvernin, V.; Ciufo, S.; Li, W. Prokaryotic genome annotation pipeline. In The NCBI Handbook, 2nd ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2013. Available online: http://www.ncbi.nlm.nih.gov/books/NBK174280 (accessed on 1 August 2023).
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, gkad326. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein families for the microbial genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I. and P. Bork, Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/genome/ (accessed on 15 November 2021).
- KEGG PATHWAY Database. Available online: https://www.genome.jp/kegg/pathway.html (accessed on 1 July 2023).
- Harrison, K.J.; de Crécy-Lagard, V.; Zallot, R. Gene Graphics: A genomic neighborhood data visualization web application. Bioinformatics 2017, 34, 1406–1408. [Google Scholar] [CrossRef]
- Andersen, S.M.; Johnsen, K.; Sørensen, J.; Nielsen, P.; Jacobsen, C.S. Pseudomonas frederiksbergensis sp. nov., isolated from soil at a coal gasification site. Int. J. Syst. Evol. Microbiol. 2000, 50, 1957–1964. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Li, W.J.; Kroppenstedt, R.M.; Kim, C.J.; Chen, G.Z.; Park, D.J.; Xu, L.H.; Jiang, C.L. Rhodococcus yunnanensis sp. nov., a mesophilic actinobacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2005, 55, 1133–1137. [Google Scholar] [CrossRef]
- Zhang, D.C.; Schumann, P.; Liu, H.C.; Xin, Y.H.; Zhou, Y.G.; Schinner, F.; Margesin, R. Arthrobacter alpinus sp. nov., a psychrophilic bacterium isolated from alpine soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 2149–2153. [Google Scholar] [CrossRef]
- Dodor, D.E.; Tabatabai, M.A. Arylamidase activity as an index of nitrogen mineralization in soils. Commun. Soil Sci. Plant Anal. 2007, 38, 2197–2207. [Google Scholar] [CrossRef]
- Margesin, R.; Miteva, V. Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 2011, 162, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 2, 971. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, A.; Ahmed, B.; Khan, M.S.; Umar, S.; Lee, J. Psychrophilic bacterial phosphate-biofertilizers: A novel extremophile for sustainable crop production under cold environment. Microorganisms 2021, 28, 2451. [Google Scholar] [CrossRef] [PubMed]
- Ruberto, L.; Vazquez, S.C.; Mac Cormack, W.P. Effectiveness of the natural bacterial flora, biostimulation, and bioaugmentation on the bioremediation of a hydrocarbon-contaminated Antarctic soil. Int. Biodeterior. Biodegrad. 2003, 52, 115–125. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Kim, J. Sphingomonas naphthae sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 4621–4627. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, W.F.; Xu, Z.L.; Li, W.; Zhang, X.Y.; Li, W.J.; Wang, L. Sphingomonas oleivorans sp. nov., isolated from oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3720–3725. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Monisha, T.R.; Ismailsab, M.; Masarbo, R.; Nayak, A.S.; Karegoudar, T.B. Degradation of cinnamic acid by a newly isolated bacterium Stenotrophomonas sp. TRMK2. 3 Biotech. 2018, 8, 368. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Kawasaki, A. Hydrocarbon-Degrading Sphingomonads: Sphingomonas, Sphingobium, Novosphingobium, and Sphingopyxis. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Oelschlagel, M.; Ruckert, C.; Kalinowski, J.; Schmidt, G.; Schlomann, M.; Tischler, D. Sphingopyxis fribergensis sp. nov., a soil bacterium with the ability to degrade styrene and phenylacetic acid. Int. J. Syst. Evol. Microbiol. 2015, 65, 3008–3015. [Google Scholar] [CrossRef]
- Whyte, L.G.; Schultz, A.; Beilen, J.B.; Luz, A.P.; Pellizari, V.; Labbé, D.; Greer, C.W. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 2002, 41, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Tiedje, J.M. Coping with our cold planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semenova, E.M.; Tourova, T.P.; Babich, T.L.; Logvinova, E.Y.; Sokolova, D.S.; Loiko, N.G.; Myazin, V.A.; Korneykova, M.V.; Mardanov, A.V.; Nazina, T.N. Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41. Microorganisms 2024, 12, 79. https://doi.org/10.3390/microorganisms12010079
Semenova EM, Tourova TP, Babich TL, Logvinova EY, Sokolova DS, Loiko NG, Myazin VA, Korneykova MV, Mardanov AV, Nazina TN. Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41. Microorganisms. 2024; 12(1):79. https://doi.org/10.3390/microorganisms12010079
Chicago/Turabian StyleSemenova, Ekaterina M., Tatyana P. Tourova, Tamara L. Babich, Ekaterina Y. Logvinova, Diyana S. Sokolova, Nataliya G. Loiko, Vladimir A. Myazin, Maria V. Korneykova, Andrey V. Mardanov, and Tamara N. Nazina. 2024. "Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41" Microorganisms 12, no. 1: 79. https://doi.org/10.3390/microorganisms12010079
APA StyleSemenova, E. M., Tourova, T. P., Babich, T. L., Logvinova, E. Y., Sokolova, D. S., Loiko, N. G., Myazin, V. A., Korneykova, M. V., Mardanov, A. V., & Nazina, T. N. (2024). Crude Oil Degradation in Temperatures Below the Freezing Point by Bacteria from Hydrocarbon-Contaminated Arctic Soils and the Genome Analysis of Sphingomonas sp. AR_OL41. Microorganisms, 12(1), 79. https://doi.org/10.3390/microorganisms12010079