Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. DNA Manipulations, Nucleotide Sequencing, and Sequence Analysis
2.3. Construction of Disruption Mutants
2.4. Growth Curves on n-Alkanes
2.5. Resting Cell Assay
2.6. RNA Isolation
2.7. Reverse Transcription (RT)-PCR and Quantitative RT-PCR (qRT-PCR)
2.8. Purification of His-Tagged alkU
2.9. Electrophoretic Mobility Shift Assays (EMSAs)
3. Results
3.1. Growth of RHA1 on n-Alkanes
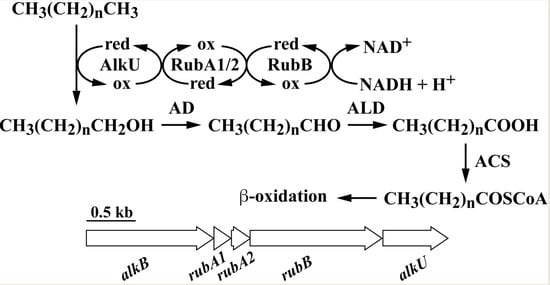
3.2. The alk Gene Cluster of Strain RHA1
3.3. Disruption of alkB in RHA1
3.4. Transcriptional Regulation of alk Operon
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 1990, 54, 305–315. [Google Scholar] [PubMed]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef] [PubMed]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, N.; Yeager, C.M.; Arp, D.J. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 2001, 67, 4992–4998. [Google Scholar] [CrossRef]
- Maier, T.; Forster, H.H.; Asperger, O.; Hahn, U. Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem. Biophys. Res. Commun. 2001, 286, 652–658. [Google Scholar] [CrossRef]
- Maeng, J.H.; Sakai, Y.; Tani, Y.; Kato, N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J. Bacteriol. 1996, 178, 3695–3700. [Google Scholar] [CrossRef]
- Watkinson, R.J.; Morgan, P. Physiology of aliphatic hydrocarbon-degrading microorganisms. Biodegradation 1990, 1, 79–92. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Panke, S.; Lucchini, S.; Franchini, A.G.; Rothlisberger, M.; Witholt, B. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: Evolution and regulation of the alk genes. Microbiology 2001, 147, 1621–1630. [Google Scholar] [CrossRef]
- Geissdörfer, W.; Kok, R.G.; Ratajczak, A.; Hellingwerf, K.J.; Hillen, W. The genes rubA and rubB for alkane degradation in Acinetobacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 1999, 181, 4292–4298. [Google Scholar]
- Ratajczak, A.; Geissdörfer, W.; Hillen, W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl. Environ. Microbiol. 1998, 64, 1175–1179. [Google Scholar]
- Ratajczak, A.; Geissdörfer, W.; Hillen, W. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 1998, 180, 5822–5827. [Google Scholar] [PubMed]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, W.; Wu, Y.; Zhou, Z.; Lai, Q.; Shao, Z. Multiple alkane hydroxylase systems in a marine alkane degrader, Alcanivorax dieselolei B-5. Environ. Microbiol. 2011, 13, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Funhoff, E.G.; Bauer, U.; Garcia-Rubio, I.; Witholt, B.; van Beilen, J.B. CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation. J. Bacteriol. 2006, 188, 5220–5227. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Sameshima, Y.; Honda, K.; Kato, J.; Omasa, T.; Ohtake, H. Expression of Rhodococcus opacus alkB genes in anhydrous organic solvents. J. Biosci. Bioeng. 2008, 106, 199–203. [Google Scholar] [CrossRef]
- Amouric, A.; Quemeneur, M.; Grossi, V.; Liebgott, P.P.; Auria, R.; Casalot, L. Identification of different alkane hydroxylase systems in Rhodococcus ruber strain SP2B, an hexane-degrading actinomycete. J. Appl. Microbiol. 2010, 108, 1903–1916. [Google Scholar] [CrossRef]
- Cappelletti, M.; Fedi, S.; Frascari, D.; Ohtake, H.; Turner, R.J.; Zannoni, D. Analyses of both the alkB gene transcriptional start site and alkB promoter-inducing properties of Rhodococcus sp. strain BCP1 grown on n-alkanes. Appl. Environ. Microbiol. 2011, 77, 1619–1627. [Google Scholar] [CrossRef]
- Takei, D.; Washio, K.; Morikawa, M. Identification of alkane hydroxylase genes in Rhodococcus sp. strain TMP2 that degrades a branched alkane. Biotechnol. Lett. 2008, 30, 1447–1452. [Google Scholar] [CrossRef]
- Whyte, L.G.; Smits, T.H.; Labbe, D.; Witholt, B.; Greer, C.W.; van Beilen, J.B. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 2002, 68, 5933–5942. [Google Scholar] [CrossRef] [PubMed]
- Lo Piccolo, L.; De Pasquale, C.; Fodale, R.; Puglia, A.M.; Quatrini, P. Involvement of an alkane hydroxylase system of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Appl. Environ. Microbiol. 2011, 77, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, P.; Scaglione, G.; De Pasquale, C.; Riela, S.; Puglia, A.M. Isolation of Gram-positive n-alkane degraders from a hydrocarbon-contaminated Mediterranean shoreline. J. Appl. Microbiol. 2008, 104, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Seto, M.; Masai, E.; Ida, M.; Hatta, T.; Kimbara, K.; Fukuda, M.; Yano, K. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 1995, 61, 4510–4513. [Google Scholar] [PubMed]
- Seto, M.; Kimbara, K.; Shimura, M.; Hatta, T.; Fukuda, M.; Yano, K. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 1995, 61, 3353–3358. [Google Scholar]
- Iwasaki, T.; Miyauchi, K.; Masai, E.; Fukuda, M. Multiple-subunit genes of the aromatic-ring-hydroxylating dioxygenase play an active role in biphenyl and polychlorinated biphenyl degradation in Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2006, 72, 5396–5402. [Google Scholar] [CrossRef]
- Mathieu, J.M.; Mohn, W.W.; Eltis, L.D.; LeBlanc, J.C.; Stewart, G.R.; Dresen, C.; Okamoto, K.; Alvarez, P.J. 7-ketocholesterol catabolism by Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2010, 76, 352–355. [Google Scholar] [CrossRef]
- Patrauchan, M.A.; Florizone, C.; Eapen, S.; Gómez-Gil, L.; Sethuraman, B.; Fukuda, M.; Davies, J.; Mohn, W.W.; Eltis, L.D. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J. Bacteriol. 2008, 190, 37–47. [Google Scholar] [CrossRef]
- Sharp, J.O.; Sales, C.M.; LeBlanc, J.C.; Liu, J.; Wood, T.K.; Eltis, L.D.; Mohn, W.W.; Alvarez-Cohen, L. An inducible propane monooxygenase is responsible for N-nitrosodimethylamine degradation by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2007, 73, 6930–6938. [Google Scholar] [CrossRef]
- Araki, N.; Niikura, Y.; Miyauchi, K.; Kasai, D.; Masai, E.; Fukuda, M. Glucose-mediated transcriptional repression of PCB/biphenyl catabolic genes in Rhodococcus jostii RHA1. J. Mol. Microb. Biotech. 2011, 20, 53–62. [Google Scholar] [CrossRef]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Short, J.M.; Fernandez, J.M.; Sorge, J.A.; Huse, W.D. λ ZAP: A bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988, 16, 7583–7600. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Nishiyama, M.; Yu, F.; Watanabe, I.; Horinouchi, S.; Beppu, T. Development of a host-vector system in a Rhodococcus strain and its use for expression of the cloned nitrile hydratase gene cluster. J. Gen. Microbiol. 1992, 138, 1003–1010. [Google Scholar] [CrossRef]
- Masai, E.; Yamada, A.; Healy, J.M.; Hatta, T.; Kimbara, K.; Fukuda, M.; Yano, K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 1995, 61, 2079–2085. [Google Scholar]
- Kasai, D.; Fujinami, T.; Abe, T.; Mase, K.; Katayama, Y.; Fukuda, M.; Masai, E. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009, 191, 6758–6768. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001, 205, 197–202. [Google Scholar] [CrossRef]
- Goncalves, E.R.; Hara, H.; Miyazawa, D.; Davies, J.E.; Eltis, L.D.; Mohn, W.W. Transcriptomic assessment of isozymes in the biphenyl pathway of Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2006, 72, 6183–6193. [Google Scholar] [CrossRef]
- Sleight, S.C.; Bartley, B.A.; Lieviant, J.A.; Sauro, H.M. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res. 2010, 38, 2624–2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamimura, N.; Takamura, K.; Hara, H.; Kasai, D.; Natsume, R.; Senda, T.; Katayama, Y.; Fukuda, M.; Masai, E. Regulatory system of the protocatechuate 4,5-cleavage pathway genes essential for lignin downstream catabolism. J. Bacteriol. 2010, 192, 3394–3405. [Google Scholar] [CrossRef] [PubMed]
- van Beilen, J.B.; Neuenschwander, M.; Smits, T.H.; Roth, C.; Balada, S.B.; Witholt, B. Rubredoxins involved in alkane oxidation. J. Bacteriol 2002, 184, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Shanklin, J.; Achim, C.; Schmidt, H.; Fox, B.G.; Münck, E. Mössbauer studies of alkane omega-hydroxylase: Evidence for a diiron cluster in an integral-membrane enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 2981–2986. [Google Scholar] [CrossRef]
- Shanklin, J.; Whittle, E.; Fox, B.G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 1994, 33, 12787–12794. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Penninga, D.; Witholt, B. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 1992, 267, 9194–9201. [Google Scholar]
- van Beilen, J.B.; Smits, T.H.; Roos, F.F.; Brunner, T.; Balada, S.B.; Rothlisberger, M.; Witholt, B. Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 2005, 187, 85–91. [Google Scholar] [CrossRef]
- Tani, A.; Ishige, T.; Sakai, Y.; Kato, N. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 2001, 183, 1819–1823. [Google Scholar] [CrossRef]
- Canosa, I.; Yuste, L.; Rojo, F. Role of the alternative sigma factor σS in expression of the AlkS regulator of the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 1999, 181, 1748–1754. [Google Scholar]
- Canosa, I.; Sanchez-Romero, J.M.; Yuste, L.; Rojo, F. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Mol. Microbiol. 2000, 35, 791–799. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Pompeani, A.J.; Irgon, J.J.; Berger, M.F.; Bulyk, M.L.; Wingreen, N.S.; Bassler, B.L. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 2008, 70, 76–88. [Google Scholar] [CrossRef]
- George, A.M.; Levy, S.B. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 1983, 155, 541–548. [Google Scholar]
- Orth, P.; Schnappinger, D.; Hillen, W.; Saenger, W.; Hinrichs, W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000, 7, 215–219. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Nodwell, J.R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 2013, 77, 440–475. [Google Scholar] [CrossRef]







| Strain or Plasmid | Relevant Characteristic(s) a | Source or Reference |
|---|---|---|
| Strains | ||
| R. jostii | ||
| RHA1 | Wild type | [25] |
| DAB | RHA1 derivative; ΔalkB | This study |
| DAU | RHA1 derivative; ΔalkU | This study |
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi hsdR17 relA1 supE44 Δ(lac-proAB) mcrA [F’, traD36 proAB+ laclq ΔZM15] | [31] |
| S17-1 | RK2 tra regulon, λ pir, host for pir-dependent plasmids | [32] |
| BL21(DE3) | F− ompT hsdSB(rB-mB-) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | [33] |
| Plasmids | ||
| pT7Blue | Cloning vector; T7 promoter, Ap r | Novagen |
| pBluescript II KS(+) | Cloning vector; Ap r | [34] |
| pK19mobsacB | oriT sacB Km r | [35] |
| pK4 | Rhodococcus-E. coli shuttle vector, Km r | [36] |
| pET16b | Expression vector, N-terminal His10 tag, Ap r T7 promoter | Novagen |
| pTALKBL | pT7Blue with a 917-bp PCR fragment generated by alkBL_F/R primer pair | This study |
| pTALKBR | pT7Blue with a 723-bp PCR fragment generated by alkBR_F/R primer pair | This study |
| pTALKBLR | pTALKBR with a 733-bp EcoRI-SacI fragment of pTALKBL | This study |
| pABSmobsacB | pK19mobsacB with a 1.5-kb EcoRI-HindIII fragment of pTALKBLR | This study |
| pTALKBD | pT7Blue with a 967-bp PCR fragment generated by alkBD_F/R primer pair | This study |
| pT7BL | pT7Blue with a 0.8-kb EcoRI-BamHI fragment of pTALKBL | This study |
| pT7B | pT7BL with a 0.7-kb BamHI-HindIII fragment of pTALKBD | This study |
| pK4ALKB | pK4 with a 1.5-kb EcoRI fragment of pT7B | This study |
| pTUPU | pT7Blue with a 761-bp PCR fragment generated by UPalkU_F/R primer pair | This study |
| pTDWU | pT7Blue with a 792-bp PCR fragment generated by DWalkU_F/R primer pair | This study |
| pKU | pK19mobsacB with a 1.6-kb HindIII fragment carrying part of alkU | This study |
| pETALKU | pET16b with a 0.7-kb PCR amplified fragment carrying of alkU | This study |
| Primer | Sequence (5′ to 3′) a |
|---|---|
| alkB_F | TGACGACGTCGAATATCAGC |
| alkB_R | CCTGAATGATCAGGAACGG |
| alkBrubB_F | GAGCATTCACAACGATGTGC |
| alkBrubB_R | ACAGGAAGTCCTTCGACACC |
| rubA2B_F | GTACCGATTTCAAGCTCTACC |
| rubA2B_R | CACATCCGATGAGACCTCC |
| rubBalkU_F | ACGGTCGAAGTTGGAGTGC |
| rubBalkU_R | CTTTGTAGATCGTCTGCCTGC |
| alkU_F | CGAGCGACAGAGTCGATACC |
| alkU_R | ATGAAACTCAAGGCGAGCC |
| alkBL_F | ACAGGTGAAGCTACCAGCG |
| alkBL_R | GTGAGAGCTTCTGGACTTTCC |
| alkBR_F | CTTCGGTGAGAGCTTCTGG |
| alkBR_R | AAGCTTGTCGTCGGGCACATCG (HindIII) |
| alkBD_F | AACAGCTCGAGAACGACAGG |
| alkBD_R | GAATTCCATCACCGAACTCCGC (EcoRI) |
| UPalkU_F | AAGCTTTGTCCTCGCTCGACGTGAGC (HindIII) |
| UPalkU_R | GGATCCATTGTCGGCCAGGGACGTTCG (BamHI) |
| DWalkU_F | GGATCCAACGGTCGTGGGTGAACTCG (BamHI) |
| DWalkU_R | AAGCTTAGGAACTGATCTACGCCAACC (HindIII) |
| HISalkU_F | TCGAAGGTCGTCATAGCAGACGACCGCACCGCAGACGACCGCACC |
| HISalkU_R | GGATCCTCGAGCATAAAGTACTCACGGGTGAAGTACTCACGGGTG |
| Media | Strains | n-Alkanes a | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 | C13 | C14 | C15 | C16 | C17 | C18 | C19 | C20 | C30 | ||
| Liquid | RHA1 | ND | ND | ND | ND | − | − | + | + | + | + | + | + | + | + | + | − | − |
| DAB | ND | ND | ND | ND | ND | − | − | ± | + | + | + | + | + | + | + | − | − | |
| Solid | RHA1 | − | − | − | − | − | + | + | + | + | ND | ND | ND | ND | ND | ND | ND | ND |
| DAB | ND | ND | ND | ND | ND | ± | − | ± | + | ND | ND | ND | ND | ND | ND | ND | ND | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibu, N.; Kasai, D.; Ikawa, T.; Akiyama, E.; Fukuda, M. Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms 2019, 7, 479. https://doi.org/10.3390/microorganisms7110479
Gibu N, Kasai D, Ikawa T, Akiyama E, Fukuda M. Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms. 2019; 7(11):479. https://doi.org/10.3390/microorganisms7110479
Chicago/Turabian StyleGibu, Namiko, Daisuke Kasai, Takumi Ikawa, Emiko Akiyama, and Masao Fukuda. 2019. "Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1" Microorganisms 7, no. 11: 479. https://doi.org/10.3390/microorganisms7110479
APA StyleGibu, N., Kasai, D., Ikawa, T., Akiyama, E., & Fukuda, M. (2019). Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms, 7(11), 479. https://doi.org/10.3390/microorganisms7110479