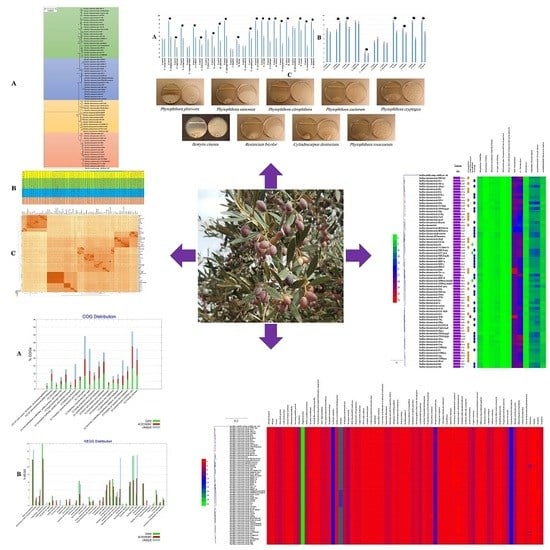
Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial, Oomycete and Fungal Strains and Growth Conditions
2.2. Plant Material
2.3. Isolation of Endophytic Bacteria and Screening for Antifungal Activity
2.4. Antifungal Activity of B. velezensis OEE1 against Oomycetes and True Fungi
2.5. Inhibition of F. solani Fso1 Mycelial Growth by Extracellular B. velezensis OEE1 Metabolites
2.6. Effect of Extracellular B. velezensis OEE1 Metabolites on F. solani Fso1 Conidia Germination
2.7. Antifungal Traits of B. velezensis Strain OEE1
2.7.1. Detection of Protease Production
2.7.2. Chitinase and β-glucanase Activity
2.7.3. Hydrogen Cyanide (HCN) Production
2.8. Plant Growth Promotion (PGP) Traits of the B. velezensis Strain OEE1
2.8.1. Siderophore Production
2.8.2. Production of Indole-3-Acetic Acid (IAA)
2.8.3. Phosphate Solubilisation
2.8.4. Nitrogen Fixation
2.9. Endophytic Traits of B. velezensis Strain OEE1
2.9.1. Cellulase Activity
2.9.2. Pectinase Activity
2.9.3. Amylase Activity
2.10. Samples Preparation for GC-MS and LC-HRMS Analysis
2.11. GC-MS Analysis
2.12. LC-HRMS Analysis: LC-HRMS Instrumentation and Conditions
2.13. Bacterial DNA Extraction and Amplification
2.14. Bacterial Genome Sequencing Assembly and Annotation
2.15. Selection of Genomes for Phylogenomic Analysis
2.16. Whole Genome Phylogeny
2.17. Average Nucleotide Identity (ANI) Analysis
2.18. Genome-to-Genome Distance Calculator (GGDC) Analysis
2.19. Comparative Genomics Analysis
2.20. Identification of Core Genome and Accessory Genomes of the Isolate Collection
2.21. B. velezensis Strain OEE1 Application and F. solani Infection of Olive Trees under Greenhouse Conditions
2.21.1. Biocontrol Assay
2.21.2. Plant Growth Measurement
2.21.3. Colonization Assays
2.22. Statistical Analysis
3. Results
3.1. Isolation of Endophytic Bacteria and Screening for Antifungal Activity
3.2. Inhibition of Plant Pathogens by the B. velezensis Strain OEE1
3.3. Antifungal Traits of B. velezensis Strain OEE1
3.4. B. velezensis Strain OEE1 Plant Growth Promoting Traits
3.5. Bacillus velezensis Strain OEE1 Endophytic Traits
3.6. Phylogenetic Affinities of B. velezensis Strain OEE1
3.7. GC-MS Analysis of the B. velezensis Strain OEE1
3.8. LC-HRMS Analysis of B. velezensis Strain OEE1
3.9. Bioinformatic Evaluation of PGP Potential of B. velezensis Strains
3.10. Secondary Metabolite Clusters
3.11. Bacillus Velezensis Genomes Prediction of Natural Products Richness and Location
3.12. Characterization of B. velezensis Core and Pan Genomes
3.13. Functional Characterization of the Core, Accessory and Unique Genomes of B. velezensis
3.14. Application of B. velezensis Strain OEE1 under Greenhouse Conditions
3.15. Endophytic Colonization of B. velezensis Strain OEE1 of Olive Trees Cultivar Chemlali Roots
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Belbahri, L.; Calmin, G.; Mauch, F.; Andersson, J.O. Functional and evolutionary studies of Phytophthora brassicae cutinase: Evidence for lateral gene transfer of a crucial oomycete virulence factor. Gene 2008, 408, 1–8. [Google Scholar] [CrossRef]
- Lara, E.; Belbahri, L. SSU rRNA reveals major trends in oomycete evolution. Fungal Divers. 2011, 49, 93–100. [Google Scholar] [CrossRef]
- De Vita, P.; Serrano, M.S.; Belbahri, L.; Garcia, L.V.; Ramo, C.; Sanchez, M.E. Germination of hyphal bodies of Pythium spiculum isolated from declining cork oaks at Donana National Park (Spain). Phytopathol. Mediterr. 2011, 50, 478–481. [Google Scholar]
- Prospero, S.; Vercauteren, A.; Heungens, K.; Belbahri, L.; Rigling, D. Phytophthora diversity and the population structure of Phytophthora ramorum in Swiss ornamental nurseries. Plant Pathol. 2013, 62, 1063–1071. [Google Scholar] [CrossRef]
- Steciow, M.M.; Lara, E.; Pillonel, A.; Pelizza, S.A.; Lestani, E.A.; Rossi, G.C.; Belbahri, L. Incipient loss of flagella in the genus Geolegnia: The emergence of a new clade within Leptolegnia? IMA Fungus 2013, 4, 169–175. [Google Scholar] [CrossRef]
- Steciow, M.M.; Lara, E.; Paul, C.; Pillonel, A.; Belbahri, L. Multiple barcode assessment within the Saprolegnia-Achlya clade (Saprolegniales, Oomycota, Straminipila) brings order in a neglected group of pathogens. IMA Fungus 2014, 5, 439–448. [Google Scholar] [CrossRef]
- Moralejo, E.; Perez-Sierra, A.; Alvarez, L.; Belbahri, L.; Lefort, F.; Descals, E. Multiple alien Phytophthoras discovered on diseased ornamental plants in Spain. Plant Pathol. 2009, 58, 100–110. [Google Scholar] [CrossRef]
- Abad, Z.G.; Abad, J.A.; Cunnington, J.H.; Smith, I.W.; Blomquist, C.; Balci, Y.; Moralejo, E.; Pérez-Sierra, A.; Abad-Campos, P.; Alvarez-Bernaola, L.A.; et al. Phytophthora niederhauserii sp. nov., a new polyphagous species mostly isolated from ornamentals potted plants in twelve countries of five continents. Mycologia 2014, 106, 431–447. [Google Scholar] [CrossRef]
- Belbahri, L.; Moralejo, E.; Calmin, G.; Oszako, T.; Garcia, J.A.; Descals, E.; Lefort, F. Phytophthora polonica, a new species isolated from declining Alnus glutinosa stands in Poland. FEMS Microbiol. Lett. 2006, 261, 165–174. [Google Scholar] [CrossRef]
- Chavarriaga, D.; Bodles, W.J.A.; Leifert, C.; Belbahri, L.; Woodward, S. Phytophthora cinnamomi and other fine root pathogens in north temperate pine forests. FEMS Microbiol. Lett. 2007, 276, 67–74. [Google Scholar] [CrossRef]
- Belbahri, L.; Calmin, G.; Wagner, S.; Moralejo, E.; Woodward, S.; Lefort, F. Specific hybridization real-time PCR probes for Phytophthora ramorum detection and diagnosis. For. Pathol. 2007, 37, 403–408. [Google Scholar] [CrossRef]
- Romero, M.A.; Sanchez, J.E.; Jimenez, J.J.; Belbahri, L.; Trapero, A.; Lefort, F.; Sanchez, M.E. New Pythium taxa causing root rot on Mediterranean Quercus species in SW Spain and Portugal. J. Phytopathol. 2007, 155, 289–295. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Fraser, S.; Belka, M.; Dogmuş, T.H.; Heckova, Z.; Oskay, F.; Belbahri, L.; Woodward, S. Biological control of Dothistroma needle blight on pine with Aneurinibacillus migulanus. For. Pathol. 2016, 46, 555–558. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Rekik, I.; Bouket, A.C.; Luptakova, L.; Weitz, H.J.; Rateb, M.E.; Jaspars, M.; Woodward, S.; Belbahri, L. Increased biological activity of Aneurinibacillus migulanus strains correlates with the production of new gramicidin secondary metabolites. Front. Microbiol. 2017, 8, 517. [Google Scholar] [CrossRef]
- Cherrad, S.; Charnay, A.; Hernandez, C.; Steva, H.; Belbahri, L.; Vacher, S. Emergence of boscalid-resistant strains of Erysiphe necator in French vineyards. Microbiol. Res. 2018, 216, 79–84. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Rekik, I.; Bełka, M.; Ibrahim, A.F.; Luptáková, L.; Jaspars, M.; Woodward, S.; Belbahri, L. Strain-level diversity of secondary metabolism in the biocontrol species Aneurinibacillus migulanus. Microbiol. Res. 2016, 182, 116–124. [Google Scholar] [CrossRef]
- Ben Slama, H.; Cherif-Silini, H.; Bouket, A.C.; Qader, M.; Silini, A.; Yahiaoui, B.; Alenezi, F.N.; Luptakova, L.; Triki, M.A.; Vallat, A.; et al. Screening for Fusarium antagonistic bacteria from contrasting niches designated the endophyte Bacillus halotolerans as plant warden against Fusarium. Front. Microbiol. 2019, 9, 3236. [Google Scholar] [CrossRef]
- Belbahri, L.; Alenezi, F.N.; Luptáková, L.; Rateb, M.E.; Woodward, S. Complete genome sequence of Aneurinibacillus migulanus E1, a Gramicidin S- and d-Phenylalanyl-l-Propyl Diketopiperazine-deficient mutant. Genome Announc. 2015, 3, e01441-15. [Google Scholar] [CrossRef]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Oszako, T.; Cherrad, S.; Vacher, S.; Rateb, M.E. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for adaptation to plant-associated habitats and a dynamic accessory genome with wealth of unsuspected new and unlocked secondary metabolites arsenal. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Mefteh, F.B.; Daoud, A.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Rateb, M.E.; Kadri, A.; Gharsallah, N.; Belbahri, L. Fungal root microbiome from healthy and brittle leaf diseased date palm trees (Phoenix dactylifera L.) reveals a hidden untapped arsenal of antibacterial and broad spectrum antifungal secondary metabolites. Front. Microbiol. 2017, 8, 307. [Google Scholar] [CrossRef]
- Ben Mefteh, F.; Daoud, A.; Bouket, A.C.; Thissera, B.; Kadri, Y.; Cherif-Silini, H.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Oszako, T.; et al. Date palm trees root-derived endophytes as fungal cell factories for diverse bioactive metabolites. Int. J. Mol. Sci. 2018, 19, 1986. [Google Scholar] [CrossRef]
- Mefteh, B.F.; Frikha, F.; Daoud, A.; Chenari Bouket, A.; Luptakova, L.; Alenezi, F.N.; Al-Anzi, B.S.; Oszako, T.; Gharsallah, N.; Belbahri, L. Response surface methodology optimization of an acidic protease produced by Penicillium bilaiae isolate TDPEF30, a newly recovered endophytic fungus from healthy roots of date palm trees (Phoenix dactylifera L.). Microorganisms 2019, 7, 74. [Google Scholar] [CrossRef]
- Zouari, I.; Jlaiel, L.; Tounsi, S.; Trigui, M. Biocontrol activity of the endophytic Bacillus amyloliquefaciens strain CEIZ-11 against Pythium aphanidermatum and purification of its bioactive compounds. Biol. Control 2016, 100, 54–62. [Google Scholar] [CrossRef]
- Khlifi, R.; Belbahri, L.; Woodward, S.; Mechichi, T. Decolourization and detoxification of textile industry wastewater by the laccase-mediator system. J. Hazard. Mater. 2009, 175, 802–808. [Google Scholar] [CrossRef]
- Costa, L.E.D.O.; De Queiroz, M.V.; De Moraes, C.A.; De Araújo, E.F.; Borges, A.C. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 2012, 43, 1562–1575. [Google Scholar] [CrossRef]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Weitz, H.J.; Belbahri, L.; Nidhal, J.; Luptáková, L.; Jaspars, M.; Woodward, S. Draft genome sequence of Aneurinibacillus migulanus NCTC 7096. Genome Announc. 2015, 3, e00234-15. [Google Scholar] [CrossRef]
- Alenezi, F.N.; Weitz, H.J.; Belbahri, L.; Ben Rebah, H.; Luptáková, L.; Jaspars, M.; Woodward, S. Draft genome sequence of Aneurinibacillus migulanus strain Nagano. Genome Announc. 2015, 3, e00232-15. [Google Scholar] [CrossRef]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; Van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Goris, J.; Klappenbach, J.A.; Vandamme, P.; Coenye, T.; Konstantinidis, K.T.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Ozer, E.A.; Allen, J.P.; Hauser, A.R. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genom. 2014, 15, 737. [Google Scholar] [CrossRef]
- Gharbi, Y.; Triki, M.A.; Trabelsi, R.; Fendri, I.; Daayf, F.; Gdoura, R. Genetic structure of Verticillium dahlia isolates infecting olive trees in Tunisia using AFLP, pathogenicity and PCR markers. Plant Pathol. 2014, 64, 871–879. [Google Scholar] [CrossRef]
- Trabelsi, R.; Sellami, H.; Gharbi, Y.; Cheffi, M.; Chaari, A.; Baucher, M.; El Jaziri, M.; Triki, M.A.; Gdoura, R. Response of olive tree (Olea europaea L.cv. Chemlali) to infection with soil-borne fungi. J. Plant Dis. Prot. 2017, 124, 153. [Google Scholar] [CrossRef]
- Fan, B.; Blom, J.; Klenk, H.-P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef]
- Kim, K.; Kim, N.-J.; Kim, S.Y.; Kim, I.H.; Kim, K.-S.; Lee, G.R. Cyclo(Phe-Pro) Produced by the human pathogen Vibrio vulnificus inhibits host innate immune responses through the NF-κB pathway. Infect. Immun. 2015, 83, 1150–1161. [Google Scholar] [CrossRef]
- Gao, L.; Han, J.; Liu, H.; Qu, X.; Lu, Z.; Bie, X. Plipastatin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie Leeuwenhoek 2017, 110, 1007–1018. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The Natural Product Domain Seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef]
- Skinnider, M.A.; DeJong, C.A.; Rees, P.N.; Johnston, C.W.; Li, H.; Webster, A.L.H.; Wyatt, M.A.; Magarvey, N.A. Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res. 2015, 43, 9645–9662. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal post translationally modified peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Li, M.H.; Ung, P.M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D.H. Automated genome mining for natural products. BMC Bioinform. 2009, 10, 185. [Google Scholar] [CrossRef]
- Hocking, M.B. Handbook of Chemical Technology and Pollution Control, 3rd ed.; Academic Press: Cambridge, MA, USA, 2005; p. 830. [Google Scholar] [CrossRef]
- Lingappa, B.T.; Prasad, M.; Lingappa, Y.; Hunt, D.F.; Biemann, K. Phenethyl alcohol and Tryptophol: Autoantibiotics produced by the fungus Candida albicans. Science 1969, 163, 192–194. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Ferracane, J.L. Materials in Dentistry: Principles and Applications, 2nd ed.; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2001. [Google Scholar]
- Ghelardini, C.; Galeotti, N.; Mannelli, L.D.C.; Mazzanti, G.; Bartolini, A. Local anaesthetic activity of β-caryophyllene. Farmaco 2001, 56, 387–389. [Google Scholar] [CrossRef]
- Attokaran, M. Natural Food Flavors and Colorants; John Wiley and Sons: New York, NY, USA, 2011. [Google Scholar]
- Kumar, S.N.; Mohandas, C.; Nambisan, B. Purification of an antifungal compound, cyclo(l-Pro-d-Leu) for cereals produced by Bacillus cereus subsp. thuringiensis associated with entomopathogenic nematode. Microbiol. Res. 2013, 168, 278–288. [Google Scholar] [CrossRef]
- Andrade, J.; Ramírez, R.; Conde, M.; Sobrino, F.; Bedoya, F.J. Nicotinamide inhibits inducible nitric oxide synthase enzyme activity in macrophages by allowing nitric oxide to inhibit its own formation. Life Sci. 1997, 61, 1843–1850. [Google Scholar] [CrossRef]
- Keeling, C.I.; Slessor, K.N.; Higo, H.A.; Winston, M.L. New components of the honey bee (Apis mellifera L.) queen retinue pheromone. Proc. Natl. Acad. Sci. USA 2003, 100, 4486–4491. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K.S.M. Bioemulsifiers are not biosurfactants and require different screening approaches. Front. Microbiol. 2015, 6, 245. [Google Scholar] [CrossRef]
- Pan, J.; Hu, C.; Yu, J.-H. Lipid biosynthesis as an antifungal target. J. Fungi 2018, 4, 50. [Google Scholar] [CrossRef]
- Severino, V.; Farina, A.; Fleischmann, F.; Dalio, R.J.D.; Di Maro, A.; Scognamiglio, M.; Fiorentino, A.; Parente, A.; Osswald, W.; Chambery, A. Molecular profiling of the Phytophthora plurivora secretome: A step towards understanding the cross-talk between plant pathogenic oomycetes and their hosts. PLoS ONE 2014, 9, e112317. [Google Scholar] [CrossRef]
- Montes, M.S.; Nielsen, B.J.; Schmidt, S.G.; Bodker, L.; Kjoller, R.; Rosendahl, S. Population genetics of Phytophthora infestans in Denmark reveals dominantly clonal populations and specific alleles linked to Metalaxyl-M resistance. Plant Pathol. 2016, 65, 744–753. [Google Scholar] [CrossRef]
- Di Francesco, A.; Milella, F.; Mari, M.; Roberti, R. A preliminary investigation in to Aureobasidium pullulans as a potential biocontrol agent against Phytophthora infestans of tomato. Biol. Control 2017, 114, 144–149. [Google Scholar] [CrossRef]
- Widmer, T.L.; Shishkoff, N. Reducing infection and secondary inoculum of Phytophthora ramorum on Viburnum tinus roots grown in potting medium amended with Trichoderma asperellum isolate 04–22. Biol. Control 2017, 107, 60–69. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; Henson, J.; Van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.-O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.J.; White, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Kurbetli, I.; Ozan, S. Occurrence of Phytophthora Root and Stem Rot of Kiwifruit in Turkey. J. Phytopathol. 2013, 161, 887–889. [Google Scholar] [CrossRef]
- Gonzalez, M.; Pérez-Sierra, A.; Serrano, M.S.; Sanchez, M.E. Two Phytophthora species causing decline of wild olive (Olea europaea subsp. europaea var. sylvestris). Plant Pathol. 2017, 66, 941–948. [Google Scholar]
- Lozano-Tovar, M.; Garrido-Jurado, I.; Quesada-Moraga, E.; Raya-Ortega, M.; Trapero-Casas, A. Metarhizium brunneum and Beauveria bassiana release secondary metabolites with antagonistic activity against Verticillium dahliae and Phytophthora megasperma olive pathogens. Crop. Prot. 2017, 100, 186–195. [Google Scholar] [CrossRef]
- Bailly, A.; Weisskopf, L. Mining the volatilomes of plant-associated microbiota for new biocontrol solutions. Front. Microbiol. 2017, 8, 1638. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Safdar, A.; Huang, Z.; Rajer, F.U.; Gao, X. Effect of volatile compounds produced by Ralstonia solanacearum on plant growth promoting and systemic resistance inducing potential of Bacillus volatiles. BMC Plant Biol. 2017, 17, 133. [Google Scholar] [CrossRef]
- Sánchez-López, Á.M.; Baslam, M.; De Diego, N.; Muñoz, F.J.; Bahaji, A.; Almagro, G.; Ricarte-Bermejo, A.; García-Gómez, P.; Li, J.; Humplík, J.F.; et al. Volatile compounds emitted by diverse phytopathogenic microorganisms promote plant growth and flowering through cytokinin action. Plant Cell Environ. 2016, 39, 2592–2608. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Ruan, W.; Li, J.; Xu, H.; Wang, J.; Gao, Y.; Wang, J. Biological control of phytopathogenic fungi by fatty acids. Mycopathologia 2008, 166, 93–102. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.-J.; Kwon, S.-W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W.; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef]









| Retention time (Rt) | Kovat’s Index | Lib Score | Tentative Identification | Plant Growth Promotion (PGP) Effectiveness or Biocontrol Ability | Reference |
|---|---|---|---|---|---|
| 8.981 | 854 | 92.0 | Ethylbenzene | Used as pesticides | [50] |
| 11.473 | 1082 | 93.8 | Phenylethyl Alcohol | Antimicrobial activity, an auto-antibiotic produced by the fungus Candida albicans | [51] |
| 13.909 | 1174 | 84.6 | Indole | As an intercellular signal molecule, indole regulates various aspects of bacterial physiology, including spore formation, plasmid stability, resistance to drugs, biofilm formation and virulence | [52] |
| 16.368 | 1187 | 94.5 | 1-Dodecene | ||
| 18.656 | 1251 | 92.1 | Benzene acetic acid (Phenylacetic acid) | An active auxin (a type of plant hormone), naturally produced by the metapleural gland of most ant species and used as an antimicrobial | |
| 20.942 | 1337 | 82.1 | Eugenol | Zinc oxide eugenol is used for root canal sealing | [53] |
| 21.103 | 1424 | 69.8 | E-Caryophyllene | Widely distributed among plant oils, and reportedly possess acaricidal, insecticidal, repellent, attractive and antifungal properties | [54] |
| 22.116 | 1587 | 93.9 | 1-Hexadecene | ||
| 22.995 | 1478 | 82.1 | β-Selinene (β-Eudesmene) | A sesquiterpene hydrocarbon with a naphthalene skeleton and is a component of celery oil. The autoxidation product of it has antimalarial activity | [55] |
| 23.563 | 1496 | 79.8 | Cyclo(Leu-Pro) | Strongly inhibit mycelia growth of fungus and thereby affecting aflatoxin production | [56] |
| 24.498 | 1627 | 71.2 | 3-Hydroxy-β-damascone | Inducer of NAD(P)H:quinone reductase (QR) activity; novel inhibitors of inducible nitric oxide synthase (iNOS) induction | [57] |
| Rt | Kovat’s Index | Lib Score | Tentative Identification | PGP Effectiveness or Biocontrol Ability | Reference |
| 25.458 | 1864 | 78.5 | N-cetyl alcohol | ||
| 25.897 | 1891 | 92.1 | 1-Nonadecene | ||
| 26.172 | 1908 | 72.5 | Palmitic acid methyl ester | ||
| 30.796 | 1942 | 83.2 | Palmitic acid | ||
| 31.173 | 2070 | 79.3 | 1-Octadecanol (Stearyl alcohol) | ||
| 31.395 | 2073 | 93.6 | Methyl linoleate | ||
| 32.924 | 2084 | 67.1 | Methyl oleate | Identified as a primer pheromone in honeybees | [58] |
| 33.687 | 2138 | 94.1 | Cyclo(Phe-Pro) | A secondary metabolite produced by certain bacteria and fungi controls the expression of genes involved in pathogenicity, cell-to-cell communication by bacteria, It also modulates auxin signaling in plants. | [59] |
| 39.015 | 2704 | 73.5 | Diisooctyl phthalate |
| Retention time (Rt) | Tentative Identification | Formula of the Molecule | PGP Effectiveness or Biocontrol Ability | Reference |
|---|---|---|---|---|
| 22.06 | Surfactin B | C52H91N7O13 | A heptapeptide linked with a C13–C16 β-OH hydroxyl fatty acid, is a type of biosurfactant with antibacterial and haemolytic activities, but it has no inhibitory effects on filamentous fungi. | [61] |
| 22.73 | Surfactin C15 | C53H93N7O13 | ||
| 14.88 | Plipastatin B1 | C74H114N12O20 | Composed of ten amino acids linked to a C14–C18 β-OH fatty acid, is an antifungal metabolite that inhibits filamentous fungi, while it has no effect on yeasts and bacteria. | [62] |
| 14.12 | C16-Fengycin B | C73H112N12O20 | Composed of ten amino acids linked to a C14–C18 β-OH fatty acid, is an antifungal metabolite that inhibits filamentous fungi, while it has no effect on yeasts and bacteria. | [62] |
| 14.41 | Fengycin IX | C72H110N12O20 | ||
| 15.41 | Fengycin XII | C75H116N12O20 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheffi, M.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms 2019, 7, 314. https://doi.org/10.3390/microorganisms7090314
Cheffi M, Chenari Bouket A, Alenezi FN, Luptakova L, Belka M, Vallat A, Rateb ME, Tounsi S, Triki MA, Belbahri L. Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms. 2019; 7(9):314. https://doi.org/10.3390/microorganisms7090314
Chicago/Turabian StyleCheffi, Manel, Ali Chenari Bouket, Faizah N. Alenezi, Lenka Luptakova, Marta Belka, Armelle Vallat, Mostafa E. Rateb, Slim Tounsi, Mohamed Ali Triki, and Lassaad Belbahri. 2019. "Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes" Microorganisms 7, no. 9: 314. https://doi.org/10.3390/microorganisms7090314
APA StyleCheffi, M., Chenari Bouket, A., Alenezi, F. N., Luptakova, L., Belka, M., Vallat, A., Rateb, M. E., Tounsi, S., Triki, M. A., & Belbahri, L. (2019). Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms, 7(9), 314. https://doi.org/10.3390/microorganisms7090314