Genomic and Metabolic Insights into Two Novel Thiothrix Species from Enhanced Biological Phosphorus Removal Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. PAO-Enriched Laboratory Culture for Thiothrix sp. RT
2.2. Metagenome Sequencing and Assembly of Thiothrix sp. RT
2.3. Genome of Thiothrix sp. SSD2
2.4. Phylogenetic Analysis of MAGs
2.5. Nucleotide Sequence Accession Number
3. Results
3.1. Assembly of the Genome of New Member of the Genus Thiothrix from EBPR Bioreactor
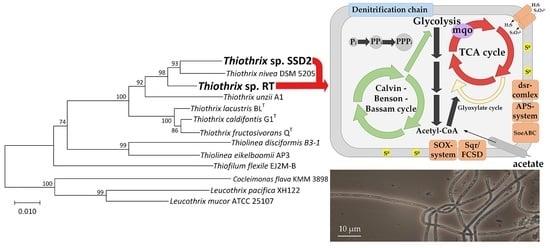
3.2. Phylogenetic Analysis of MAGs
3.3. Comparative Analysis of Metabolic Pathways
3.3.1. Dissimilatory Metabolism of Sulfur Compounds
3.3.2. Autotrophic CO2 Assimilation
3.3.3. Assimilation of N2
3.3.4. Dissimilatory Nitrate Reduction
3.3.5. Phosphorus Accumulation
3.3.6. Central Metabolic Pathways
3.3.7. Respiratory Electron Transport Chain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Larkin, J.M.; Shinabarger, D.L. Characterization of Thiothrix nivea. Int. J. Syst. Bacteriol. 1983, 33, 841–846. [Google Scholar] [CrossRef] [Green Version]
- Odintsova, E.V.; Wood, A.P.; Kelly, D.P. Chemolithoautotrophic growth of Thiothrix ramose. Arch. Microbiol. 1993, 160, 152–157. [Google Scholar] [CrossRef]
- Unz, R.F.; Head, I.M. Thiothrix. In Bergey’s Manual of Systematic Bacteriology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–16. [Google Scholar] [CrossRef]
- Boden, R.; Scott, K.M. Evaluation of the genus Thiothrix Winogradsky 1888 (Approved Lists 1980) emend. Aruga et al. 2002: Reclassification of Thiothrix disciformis to Thiolinea disciformis gen. nov., comb. nov., and of Thiothrix flexilis to Thiofilum flexile gen. nov., comb nov., with emended description of Thiothrix. Int. J. Syst. Evol. Microbiol. 2018, 68, 2226–2239. [Google Scholar] [CrossRef] [PubMed]
- Dudek, N.K.; Sun, C.L.; Burstein, D.; Kantor, R.S.; Aliaga Goltsman, D.S.; Bik, E.M.; Thomas, B.C.; Banfield, J.F.; Relman, D.A. Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome. Curr. Biol. 2017, 27, 3752–3762.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bik, E.M.; Costello, E.K.; Switzer, A.D.; Callahan, B.J.; Holmes, S.P.; Wells, R.S.; Carlin, K.P.; Jensen, E.D.; Venn-Watson, S.; Relman, D.A. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 2016, 7, 10516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharrar, A.M.; Flood, B.E.; Bailey, J.V.; Jones, D.S.; Biddanda, B.A.; Ruberg, S.A.; Marcus, D.N.; Dick, G.J. Novel large sulfur bacteria in the metagenomes of groundwater-fed chemosynthetic microbial mats in the Lake Huron basin. Front. Microbiol. 2017, 8, 791. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Pachiadaki, M.G.; Brown, J.M.; Brown, J.; Bezuidt, O.; Berube, P.M.; Biller, S.J.; Poulton, N.J.; Burkart, M.D.; La Clair, J.J.; Chisholm, S.W.; et al. Charting the complexity of the marine microbiome through single-cell genomics. Cell 2019, 179, 1623–1635. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Xu, W.; Pan, J.; Luo, Z.-H.; Li, M. Genome- and community-level interaction insights into carbon utilization and element cycling functions of Hydrothermarchaeota in hydrothermal sediment. mSystems 2020, 5, e00795-19. [Google Scholar] [CrossRef] [Green Version]
- Sears, K.; Alleman, J.E.; Barnard, J.L.; Oleszkiewicz, J.A. Impacts of reduced sulfur components on active and resting ammonia oxidizers. J. Ind. Microbiol. Biotechnol. 2004, 31, 369–378. [Google Scholar] [CrossRef]
- Lefebvre, O.; Moletta, R. Treatment of organic pollution in industrial saline wastewater: A literature review. Water Res. 2006, 40, 3671–3682. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wu, D.; Jiang, F.; Ekama, G.A.; van Loosdrecht, M.C.M.; Chen, G.-H. The demonstration of a novel sulfur cycle-based wastewater treatment process: Sulfate reduction, autotrophic denitrification, and nitrification integrated (SANI®) biological nitrogen removal process. Biotechnol. Bioeng. 2012, 109, 2778–2789. [Google Scholar] [CrossRef] [PubMed]
- Pikaar, I.; Sharma, K.R.; Hu, S.; Gernjak, W.; Keller, J.; Yuan, Z. Water engineering. Reducing sewer corrosion through integrated urban water management. Science 2014, 345, 812–814. [Google Scholar] [CrossRef] [PubMed]
- Wanner, J.; Kucman, K.; Ottová, V.; Grau, P. Effect of anaerobic conditions on activated sludge filamentous bulking in laboratory systems. Water Res. 1987, 21, 1541–1546. [Google Scholar] [CrossRef]
- Gonzalez-Gil, G.; Holliger, C. Dynamics of microbial community structure of and enhanced biological phosphorus removal by aerobic granules cultivated on propionate or acetate. Appl. Environ. Microbiol. 2011, 77, 8041–8051. [Google Scholar] [CrossRef] [Green Version]
- De Graaff, D.R.; van Loosdrecht, M.C.; Pronk, M. Biological phosphorus removal in seawater-adapted aerobic granular sludge. Water Res. 2020, 172, 115531. [Google Scholar] [CrossRef]
- Meng, Q.; Zeng, W.; Wang, B.; Fan, Z.; Peng, Y. New insights in the competition of polyphosphate-accumulating organisms and glycogen-accumulating organisms under glycogen accumulating metabolism with trace Poly-P using flow cytometry. Chem. Eng. J. 2020, 385, 123915. [Google Scholar] [CrossRef]
- Rubio-Rincón, F.J.; Welles, L.; Lopez-Vazquez, C.M.; Nierychlo, M.; Abbas, B.; Geleijnse, M.; Nielsen, P.H.; van Loosdrecht, M.C.M.; Brdjanovic, D. Long-term effects of sulphide on the enhanced biological removal of phosphorus: The symbiotic role of Thiothrix caldifontis. Water Res. 2017, 116, 53–64. [Google Scholar] [CrossRef]
- Fukushima, T.; Uda, N.; Okamoto, M.; Onuki, M.; Satoh, H.; Mino, T. Abundance of Candidatus ‘Accumulibacter phosphatis’ in enhanced biological phosphorus removal activated sludge acclimatized with different carbon sources. Microbes Environ. 2007, 22, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.; Le, V.Q.; Hansen, A.A.; Nielsen, J.L.; Nielsen, P.H. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol. Ecol. 2011, 76, 256–267. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.T.; Nielsen, J.L.; Nielsen, P.H. ‘Candidatus Halomonas Phosphatis’, a novel Polyphosphate-Accumulating Organism in Full-Scale Enhanced Biological Phosphorus Removal Plants. Environ. Microbiol. 2012, 14, 2826–2837. [Google Scholar] [CrossRef] [PubMed]
- Kotlyarov, R.Y.; Beletsky, A.V.; Ravin, N.V.; Mardanov, A.V.; Kallistova, A.Y.; Dorofeev, A.G.; Nikolaev, Y.A.; Pimenov, N.V. A Novel Phosphate-Accumulating Bacterium Identified in a Bioreactor for Phosphate Removal from Wastewater. Microbiology 2019, 88, 751–755. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: Amodular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, K.; Bessarab, I.; Haryono, M.A.S.; Liu, X.; Zuniga-Montanez, R.E.; Roy, S.; Qiu, G.; Drautz-Moses, D.I.; Law, Y.Y.; Wuertz, S.; et al. Analysis procedures for assessing recovery of high quality, complete, closed genomes from Nanopore long read metagenome sequencing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, Y.; Zheng, X. Efficient polyhydroxyalkanoates production from a waste-activated sludge alkaline fermentation liquid by activated sludge submitted to the aerobic feeding and discharge process. Environ. Sci. Technol. 2009, 43, 7734–7741. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Yarza, P.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Tamoi, M.; Ishikawa, T.; Takeda, T.; Shigeoka, S. Enzymic and molecular characterization of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC 7942: Resistance of the enzyme to hydrogen peroxide. Biochem. J. 1996, 316, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Flechner, A.; Gross, W.; Martin, W.F.; Schnarrenberger, C. Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 1999, 447, 200–202. [Google Scholar] [CrossRef]
- Vernon, S.A.; Pratte, B.S.; Thiel, T. Role of the nifB1 and nifB2 promoters in cell-type-specific expression of two Mo nitrogenases in the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 2017, 199, e00674-16. [Google Scholar] [CrossRef] [Green Version]
- Bosch, R.; Rodrıćguez-Quiñones, F.; Imperial, J. Identification of gene products from the Azotobacter vinelandii nifBfdxNnifOQ operon. FEMS Microbiol. Lett. 1997, 157, 19–25. [Google Scholar] [CrossRef]
- Trubitsyn, I.V.; Belousova, E.V.; Tutukina, M.N.; Merkel, A.Y.; Dubinina, G.A.; Grabovich, M.Y. Expansion of ability of denitrification within the filamentous colorless sulfur bacteria of the genus Thiothrix. FEMS Microbiol. Lett. 2014, 358, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio-Rincón, F.J.; Lopez-Vazquez, C.M.; Welles, L.; van Loosdrecht, M.C.M.; Brdjanovic, D. Sulphide effects on the physiology of Candidatus Accumulibacter phosphatis type I. Appl. Microbiol. Biotechnol. 2017, 101, 1661–1672. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, D.; van der Rest, M.E.; Drysch, A.; Yücel, R. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 2000, 182, 6884–6891. [Google Scholar] [CrossRef] [Green Version]
- Kretzschmar, U.; Rückert, A.; Jeoung, J.H.; Görisch, H. Malate: Quinone oxidoreductase is essential for growth on ethanol or acetate in Pseudomonas aeruginosa The GenBank accession number for the Pseudomonas aeruginosa ATCC 17933 mqo sequence reported in this work is AY129296. Microbiology 2002, 148, 3839–3847. [Google Scholar] [CrossRef] [Green Version]




| Genes of Metabolic Pathways | T. nivea DSM 5205T | T. lacustris BLT | T. caldifontis G1T | Thiothrix sp. RT | Thiothrix sp. SSD2 |
|---|---|---|---|---|---|
| Molecular nitrogen fixation | nifDKH | - | nifDKH | nifDKH | nifDKH |
| Dissimilatory nitrate reduction | - | narGHI | narGHI, nirS, cnorBC, | narGHI, cnorBC, | narGHI, nirS, cnorBC |
| Dissimilatory reduction of NO3− to NO2−, nitrate reductase from the Nap family | napAB | - | - | - | - |
| Assimilatory reduction of NO2− to NH4+ | nirBD | nirBD, | nirBD, | nirBD, | nirBD |
| Dissimilatory sulfur metabolism | soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB | soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB | soeABC, soxAXBYZ, aprAB, sat, dsr sqr, fccAB | soeABC, soxAXBZY, aprAB, sat, dsr, sqr, fccAB | soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB |
| Calvin–Benson–Bassam cycle | IAc, IAq, II, prk(2) | IAq, II, prk(2) | IAc, IAq, II, prk(2) | IAc, IAq, II, prk(2) | IAc, IAq, II, prk(2) |
| Phosphorus metabolism | phoURB, ptsSACB, ppk, epp | phoURB, ptsSACB, ppk, epp | phoURB, ptsSACB, ppk, epp | phoURB, ptsSACB, ppk, epp | phoURB, ptsSACB, ppk, epp |
| FAD-dependent malate: quinone oxidoreductase (EC 1.1.5.4) | mqo | mqo | mqo | mqo | mqo |
| Malate dehydrogenase (oxaloacetate–decarboxylating) (NADP) (EC 1.1.1.40) | maeB | maeB | maeB | maeB | maeB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mardanov, A.V.; Gruzdev, E.V.; Smolyakov, D.D.; Rudenko, T.S.; Beletsky, A.V.; Gureeva, M.V.; Markov, N.D.; Berestovskaya, Y.Y.; Pimenov, N.V.; Ravin, N.V.; et al. Genomic and Metabolic Insights into Two Novel Thiothrix Species from Enhanced Biological Phosphorus Removal Systems. Microorganisms 2020, 8, 2030. https://doi.org/10.3390/microorganisms8122030
Mardanov AV, Gruzdev EV, Smolyakov DD, Rudenko TS, Beletsky AV, Gureeva MV, Markov ND, Berestovskaya YY, Pimenov NV, Ravin NV, et al. Genomic and Metabolic Insights into Two Novel Thiothrix Species from Enhanced Biological Phosphorus Removal Systems. Microorganisms. 2020; 8(12):2030. https://doi.org/10.3390/microorganisms8122030
Chicago/Turabian StyleMardanov, Andrey V., Eugeny V. Gruzdev, Dmitry D. Smolyakov, Tatyana S. Rudenko, Alexey V. Beletsky, Maria V. Gureeva, Nikita D. Markov, Yulia Yu. Berestovskaya, Nikolai V. Pimenov, Nikolai V. Ravin, and et al. 2020. "Genomic and Metabolic Insights into Two Novel Thiothrix Species from Enhanced Biological Phosphorus Removal Systems" Microorganisms 8, no. 12: 2030. https://doi.org/10.3390/microorganisms8122030
APA StyleMardanov, A. V., Gruzdev, E. V., Smolyakov, D. D., Rudenko, T. S., Beletsky, A. V., Gureeva, M. V., Markov, N. D., Berestovskaya, Y. Y., Pimenov, N. V., Ravin, N. V., & Grabovich, M. Y. (2020). Genomic and Metabolic Insights into Two Novel Thiothrix Species from Enhanced Biological Phosphorus Removal Systems. Microorganisms, 8(12), 2030. https://doi.org/10.3390/microorganisms8122030