Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
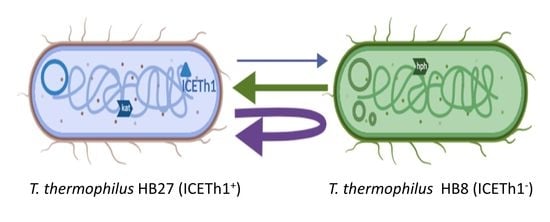
2.2. Transjugation Assays
2.3. Whole Genome Sequencing (WGS)
2.4. Parenthood Analysis
2.5. Identification of Non-Homologous Genes Acquisition
3. Results
3.1. Selection of T. thermophilus Strains for Parenthood Analysis
3.2. Transjugation between T. thermophilus HB7 and HB8 Strains
3.3. Selection of HB8-Derived Transjugants
3.4. Single Nucleotide Polymorphisms (SNPs) Reveal Parenthood of Genes
3.5. Transfer of Parental-Specific Genes
4. Discussion
4.1. Detection of Retrotransfer
4.2. Mosaicity of the Progeny
4.3. A Putative Mechanism for Retro-Transfer
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Originality-Significance Statement
References
- Garcia-Aljaro, C.; Balleste, E.; Muniesa, M. Beyond the canonical strategies of horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 2017, 38, 95–105. [Google Scholar] [CrossRef]
- Lang, A.S.; Zhaxybayeva, O.; Beatty, J.T. Gene transfer agents, phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012, 10, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Blesa, A.; Berenguer, J. Contribution of vesicle-protected extracellular DNA to horizontal gene transfer in Thermus spp. Int. Microbiol. 2015, 18, 177–187. [Google Scholar]
- Roier, S.; Zingl, F.G.; Cakar, F.; Durakovic, S.; Kohl, P.; Eichmann, T.O.; Klug, L.; Gadermaier, B.; Weinzerl, K.; Prassl, R.; et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016, 7, 10515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabezón, E.; Ripoll-Rozada, J.; Pena, A.; De la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broda, L. The formation of Hfr strains in Escherichia coli K12. Genet. Res. 1967, 9, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Hochhut, B.; Marrero, J.; Waldor, M.K. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a const in found in Vibrio cholerae O139. J. Bacteriol. 2000, 182, 2043–2047. [Google Scholar] [CrossRef] [Green Version]
- Thoma, L.; Muth, G. The conjugative DNA-transfer apparatus of Streptomyces. Int. J. Med. Microbiol. 2015, 305, 224–229. [Google Scholar] [CrossRef]
- Blesa, A.; Berenguer, J. Alternative ways to exchange DNA: Unconventional conjugation among bacteria. In Horizontal Gene Transfer; Villa, T., Viñas, M., Eds.; Springer: Cham, Germany, 2019; pp. 77–96. [Google Scholar]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef]
- Li, H. Random chromosome partitioning in the polyploid bacterium Thermus thermophilus HB27. G3 (Bethesda) 2019, 9, 1249–1261. [Google Scholar] [CrossRef] [Green Version]
- Henne, A.; Bruggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Baquedano, I.; Quintans, N.G.; Mata, C.P.; Castón, J.R.; Berenguer, J. The transjugation machinery of Thermus thermophilus, Identification of TdtA, an ATPase involved in DNA donation. PLoS Genet. 2017, 13, e1006669. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Cesar, C.E.; Averhoff, B.; Berenguer, J. Non-canonical cell-to-cell DNA transfer in Thermus spp. is insensitive to Argonaute-mediated interference. J. Bacteriol. 2015, 197, 138–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baquedano, I.; Mencia, M.; Blesa, A.; Burrus, V.; Berenguer, J. ICETh1 and ICETh2, two interdependent mobile genetic elements in Thermus thermophilus transjugation. Environ. Microbiol. 2020, 22, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Krefft, D.; Zylicz-Stachula, A.; Mulkiewicz, E.; Papkov, A.; Jezewska-Frackowiak, J.; Skowron, P.M. Two-stage gene assembly/cloning of a member of the TspDTI subfamily of bifunctional restriction endonucleases, TthHB27I. J. Biotechnol. 2015, 194, 67–80. [Google Scholar] [CrossRef]
- Baquedano, I. ICEth1 and ICEth2, Two Mobile Genetic Elements Coordinated in Thermus thermophilus Transjugation. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2019; p. 188. [Google Scholar]
- Blesa, A.; Quintans, N.G.; Baquedano, I.; Mata, C.P.; Castón, J.R.; Berenguer, J. Role of archaeal HerA protein in the biology of the bacterium Thermus thermophilus. Genes 2017, 8, 130. [Google Scholar] [CrossRef] [Green Version]
- Dordet-Frisoni, E.; Sagne, E.; Baranowski, E.; Breton, M.; Nouvel, L.X.; Blanchard, A.; Marenda, M.S.; Tardy, F.; Sirand-Pugnet, P.; Citti, C. Chromosomal transfers in mycoplasmas, when minimal genomes go mobile. mBio 2014, 5, e01958. [Google Scholar] [CrossRef] [Green Version]
- Bruggemann, H.; Chen, C. Comparative genomics of Thermus thermophilus, Plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 2006, 124, 654–661. [Google Scholar] [CrossRef]
- Cava, F.; Laptenko, O.; Borukhov, S.; Chahlafi, Z.; Blas-Galindo, E.; Gomez-Puertas, P.; Berenguer, J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 2007, 64, 630–646. [Google Scholar] [CrossRef]
- Gray, T.A.; Derbyshire, K.M. Blending genomes, distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol. Microbiol. 2018, 108, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Blesa, A.; Berenguer, J. Cell-to-cell DNA transfer among Thermus species. Bio-protocol. 2016, 6, 22006. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic, a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen, A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandmann, S.; De Graaf, A.O.; Karimi, M.; Van der Reijden, B.A.; Hellstrom-Lindberg, E.; Jansen, J.H.; Dugas, M. Evaluating variant calling tools for non-matched next-generation sequencing data. Sci. Rep. 2017, 7, 43169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit, a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff, SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos, an information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Lasa, I.; Castón, J.R.; Fernández-Herrero, L.A.; De Pedro, M.A.; Berenguer, J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol. 1992, 6, 1555–1564. [Google Scholar] [CrossRef]
- Nakamura, A.; Takakura, Y.; Kobayashi, H.; Hoshino, T. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J. Biosci. Bioeng. 2005, 100, 158–163. [Google Scholar] [CrossRef]
- Ohtani, N.; Tomita, M.; Itaya, M. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 2010, 192, 5499–5505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtani, N.; Tomita, M.; Itaya, M. The third plasmid pVV8 from Thermus thermophilus HB8, isolation, characterization, and sequence determination. Extremophiles 2012, 16, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; Sánchez, M.; Sacristán-Horcajada, E.; González-de la Fuente, S.; Peiro, R.; Berenguer, J. Into the Thermus mobilome, Presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blesa, A. Horizontal gene transfer in Thermus thermophilus: Mechanisms and barriers. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2016; p. 194. [Google Scholar]
- Dordet Frisoni, E.; Marenda, M.S.; Sagne, E.; Nouvel, L.X.; Guerillot, R.; Glaser, P.; Blanchard, A.; Tardy, F.; Sirand‐Pugnet, P.; Baranowski, E.; et al. ICEA of Mycoplasma agalactiae, a new family of self-transmissible integrative elements that confers conjugative properties to the recipient strain. Mol. Microbiol. 2013, 89, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Barany, F.; Danzitz, M.; Zebala, J.; Mayer, A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes, comparison with the isoschizomeric TaqI enzymes. Gene 1992, 112, 3–12. [Google Scholar] [CrossRef]
- Bellanger, X.; Morel, C.; Gonot, F.; Puymege, A.; Decaris, B.; Guedon, G. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol. Microbiol. 2011, 81, 912–925. [Google Scholar] [CrossRef]



| Strain | Genotype/Resistance | Use | Reference |
|---|---|---|---|
| CK1 | HB27 derivative, TTC1211(gdh1)::kat/KanR | Parental | [21] |
| CK2 | HB27 derivative, TTC1211(gdh1)::kat, ΔpilA4/KanR | Competence deficient | [22] |
| CH81 | HB8 derivative, TTHA0672::hph/HygR | Parental | This work |
| PH81 | HB8 derivative, TTHB198::hph/HygR | Parental | This work |
| Strain | % Align vs. HB27 Reference | % Align vs. HB8 Reference |
|---|---|---|
| CK1 | 98.45 | 87.53 |
| C/PH81 | 79.46 | 99.70 |
| T1 1 | 98.54 | 87.90 |
| T2 1 | 98.76 | 87.89 |
| T3 1 | 98.14 | 86.49 |
| T4 1 | 97.87 | 85.39 |
| T5 1 | 98.80 | 87.22 |
| T6 2 | 98.93 | 87.09 |
| T7 2 | 98.73 | 85.96 |
| T8 2 | 98.48 | 85.45 |
| T9 3 | 82.19 | 99.54 |
| T10 3 | 84.76 | 99.53 |
| T11 3 | 82.22 | 99.51 |
| T12 3 | 83.63 | 99.52 |
| T13 3 | 81.46 | 99.19 |
| Strain | SNPs vs. HB27 Reference | Nº of Chromosome Regions Involved 1 | SNPs vs. pTT27 Reference | Nº of pTT27 Regions Involved 1 | HB8-Specific Genes Detected |
|---|---|---|---|---|---|
| CK1 | 0 2 | - | 0 2 | - | - |
| P/CH81 | 14,818 | - | 2167 | - | - |
| T1 | 109 | 6 | 31 | 1 1 | ISTh4 |
| T2 | 6 | 6 | 283 | 5 1 | ISTh4 |
| T3 | 65 | 6 | 51 | 6 1 | TTHA0285 TTHA0286 TTHB073 |
| T4 | 92 | 6 | 0 | 1 1 | - |
| T5 | 5 | 5 | 5 | 2 1 | - |
| T6 | 39 | 5 1 | 0 | 0 | - |
| T7 | 114 | 11 1 | 0 | 0 | - |
| T8 | 50 | 2 1 | 0 | 0 | TTHA498 |
| Strain | HB8 Reference | Chromosome Regions Involved 1 | pTT27 (HB8) Reference | pTT27 Regions Involved 1 | HB27-Specific Genes Detected |
|---|---|---|---|---|---|
| CH8 | 0 | - | 0 | - | - |
| T9 | 16 | 2 1 | 0 | 0 | tdtA |
| T10 | 51 | 3 1 | 0 | 0 | TTC0952 |
| T11 | 36 | 2 1 | 0 | 0 | |
| T12 | 168 | 2 1 | 0 | 0 | TTC0398 TTC0857 |
| T13 | 14 | 2 1 | 0 | 0 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blesa, A.; Baquedano, I.; González-de la Fuente, S.; Mencía, M.; Berenguer, J. Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms 2020, 8, 2051. https://doi.org/10.3390/microorganisms8122051
Blesa A, Baquedano I, González-de la Fuente S, Mencía M, Berenguer J. Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms. 2020; 8(12):2051. https://doi.org/10.3390/microorganisms8122051
Chicago/Turabian StyleBlesa, Alba, Ignacio Baquedano, Sandra González-de la Fuente, Mario Mencía, and José Berenguer. 2020. "Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus" Microorganisms 8, no. 12: 2051. https://doi.org/10.3390/microorganisms8122051
APA StyleBlesa, A., Baquedano, I., González-de la Fuente, S., Mencía, M., & Berenguer, J. (2020). Integrative and Conjugative Element ICETh1 Functions as a Pangenomic DNA Capture Module in Thermus thermophilus. Microorganisms, 8(12), 2051. https://doi.org/10.3390/microorganisms8122051