Degradation Potential of the Nonylphenol Monooxygenase of Sphingomonas sp. NP5 for Bisphenols and Their Structural Analogs
Abstract
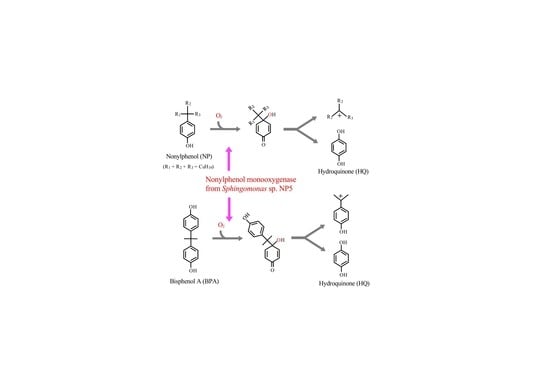
:1. Introduction
2. Materials and Methods
2.1. Chemical Compounds Used for Degradation Studies
2.2. Bacterial Strains, Plasmids, Media, and Culture Conditions Used
2.3. Degradation Tests Using Cell Suspension
2.4. HPLC and Gas Chromatography-Mass Spectrometry (GC/MS) Analyses
3. Results
3.1. Degradation of BPA and BPF
3.2. Degradation of Other BPs
3.3. Degradation of TDP and DBP
3.4. Degradation of DDE
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- United Nations Environment. Overview Report I: Worldwide Initiatives to Identify Endocrine Disrupting Chemicals (ECDs) and Potential ECDs; United Nations Environment Programme. United Nations Environment. 2017. Available online: https://wedocs.unep.org/bitstream/handle/20.500.11822/25633/EDC_report1.pdf?sequence=1&isAllowed=y (accessed on 31 January 2020).
- United Nations Environment. Overview Report II: An Overview of Current Scientific Knowledge on the Life Cycles, Environmental Exposures, and Environmental Effects of Selected Endocrine Disrupting Chemicals (EDCs) and Potential EDCs; United Nations Environment Programme. United Nations Environment. 2017. Available online: http://wedocs.unep.org/bitstream/handle/20.500.11822/25634/edc_report2.pdf?sequence=1&isAllowed=y (accessed on 31 January 2020).
- United Nations Environment. Overview Report III: Existing National, Regional, and Global Regulatory Frameworks Addressing Endocrine Disrupting Chemicals (EDCs). United Nations Environment ProgrammeUnited Nations Environment. 2017. Available online: https://wedocs.unep.org/bitstream/handle/20.500.11822/25636/edc_report3.pdf?sequence=1&isAllowed=y (accessed on 31 January 2020).
- Future Actions to Endocrine Disrupting Effects of Chemical Substances-EXTEND 2010. Ministry of the Environment, Government of Japan; 2010. Available online: https://www.env.go.jp/en/chemi/ed/extend2010_full.pdf (accessed on 31 January 2020).
- Mao, Z.; Zheng, X.-F.; Zhang, Y.-Q.; Tao, X.-X.; Li, Y.; Wang, W. Occurrence and biodegradation of nonylphenol in the environment. Int. J. Mol. Sci. 2012, 13, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Gan, J. Analysis, toxicity, occurrence and biodegradation of nonylphenol isomers: A review. Environ. Int. 2014, 73, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Careghini, A.; Mastorgio, A.F.; Saponaro, S.; Sezenna, E. Bisphenol A, nonylphenols, benzophenones, and benzotriazoles in soils, groundwater, surface water, sediments, and food: A review. Environ. Sci. Pollut. Res. 2015, 22, 5711–5741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acir, I.-H.; Guenther, K. Endocrine disrupting metabolites of aklylphenol ethoxylates-A critical review of analytical methods, environmental occurrences, toxicity, and regulation. Sci. Total Environ. 2018, 635, 1530–1546. [Google Scholar] [CrossRef] [PubMed]
- Takeo, M.; Maeda, Y.; Maeda, J.; Nishiyama, N.; Kitamura, C.; Kato, D.; Negoro, S. Two identical nonylphenol monooxygenase genes linked to IS6100 and some putative insertion sequence elements in Sphingomonas sp. NP5. Microbiology 2012, 158, 1796–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corvini, P.F.; Hollender, J.; Ji, R.; Schumacher, S.; Prell, J.; Hommes, G.; Priefer, U.; Vinken, R.; Schäffer, A. The degradation of α-quaternary nonylphenol isomers by Sphingomonas. sp. strain TTNP3 involves a type II ipso-substitution mechanism. Appl. Microbiol. Biotechnol. 2006, 70, 114–122. [Google Scholar] [CrossRef]
- Gabriel, F.L.P.; Cyris, M.; Jonkers, N.; Giger, W.; Guenther, K.; Kohler, H.-P.E. Elucidation of the ipso-substitution mechanism for side-chain cleavage of α-quaternary 4-nonylphenols and 4-t-butoxyphenol in Sphingobium xenophagum Bayram. Appl. Environ. Microbiol. 2007, 73, 3320–3326. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, F.L.P.; Cyris, M.; Giger, W.; Kohler, H.-P.E. ipso-Substitution: A general biochemical and biodegradation mechanism to cleave α-quaternary alkylphenols and bisphenol A. Chem. Biodivers. 2007, 4, 2123–2137. [Google Scholar] [CrossRef]
- Kolvenbach, B.; Schlaich, N.; Raoui, Z.; Prell, J.; Zühlke, S.; Schäffer, A.; Guengerich, F.P.; Corvini, P.F. Degradation pathway of bisphenol A: Does ipso substitution apply to phenols containing a quaternary α-carbon structure in the para position? Appl. Environ. Microbiol. 2007, 73, 4776–4784. [Google Scholar] [CrossRef] [Green Version]
- Noszczyńska, M.; Piotrowska-Seget, Z. Bisphenols: Application, occurrence, safety, and biodegradation mediated by bacterial communities in wastewater treatment plants and rivers. Chemosphere 2018, 200, 214–223. [Google Scholar] [CrossRef]
- Zhang, Z.; Alomirah, H.; Cho, H.-S.; Li, Y.-F.; Liao, C.; Minh, T.B.; Mohd, M.A.; Nakata, H.; Ren, N.; Kannan, K. Urinary bisphenol A concentrations and their implications for human exposure in several Asian countries. Environ. Sci. Technol. 2011, 45, 7044–7050. [Google Scholar] [CrossRef] [PubMed]
- Mercogliano, R.; Santonicola, S. Investigation on bisphenol A levels in human milk and daily supply chain: A review. Food Chem. Toxicol. 2018, 114, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Kannan, K.; Tan, H.; Zheng, Z.; Feng, Y.-L.; Wu, Y.; Widelka, M. Bisphenol analogues other than BPA: Environmental occurrence, human exposure, and toxicity-A review. Environ. Sci. Technol. 2016, 50, 5438–5453. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-Y.; Ike, M.; Fujita, M. Acute toxicity, mutagenicity, and estrogenicity of bisphenol-A and other bisphenols. Environ. Toxicol. 2002, 17, 80–86. [Google Scholar] [CrossRef]
- Eladak, S.; Grisin, T.; Moison, D.; Guerquin, M.J.; N’Tumba-Byn, T.; Pozzi-Gaudin, S.; Benachi, A.; Livera, G.; Rouiller-Fabre, V.; Habert, R. A new chapter in the bisphenol A story: Bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil. Steril. 2015, 103, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Rochester, J.R.; Bolden, A.L. Bisphenol S and F: A systematic review and comparison of the hormonal activity of bisphenol substitutes. Environ. Health Perspect. 2015, 123, 643–650. [Google Scholar] [CrossRef]
- Zhang, W.; Yin, K.; Chen, L. Bacteria-mediated bisphenol A degradation. Appl. Microbiol. Biotechnol. 2013, 97, 5681–5689. [Google Scholar] [CrossRef]
- Bagdasarian, M.; Lurz, R.; Rückert, B.; Franklin, F.C.; Bagdasarian, M.M.; Frey, J.; Timmis, K.N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 1981, 16, 237–247. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; CRC Press: New York, NY, USA, 2010; p. 934. [Google Scholar]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., 2nd; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Yamada, K.; Terasaki, M.; Makino, M. Estrogenic activities of alkyl(thio)phenols and 4,4′-thiodiphenol formed from degradation of commercial pesticides. J. Health Sci. 2011, 57, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Inoue, D.; Hara, S.; Kashihara, M.; Murai, Y.; Danzl, E.; Sei, K.; Tsunoi, S.; Fujita, M.; Ike, M. Degradation of bis(4-hydroxyphenyl)methane (bisphenol F) by Sphingobium yanoikuyae strain FM-2 isolated from river water. Appl. Environ. Microbiol. 2008, 74, 352–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gago-Ferrero, P.; Badia-Fabregat, M.; Olivares, A.; Piña, B.; Blánquez, P.; Vicent, T.; Caminal, G.; Díaz-Cruz, M.S.; Barceló, D. Evaluation of fungal- and photo-degradation as potential treatments for the removal of sunscreens BP3 and BP1. Sci. Total Environ. 2012, 427–428, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Mutsuga, M.; Kato, T.; Iida, M.; Tanamoto, K. Estrogenic and anti-androgenic activities of benzophenones in human estrogen and androgen receptor mediated mammalian reporter gene assay. J. Health Sci. 2005, 51, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Pijnenburg, A.M.; Everts, J.W.; de Boer, J.; Boon, J.P. Polybrominated biphenyl and diphenylether flame retardants: Analysis, toxicity, and environmental occurrence. Rev. Environ. Contam. Toxicol. 1995, 141, 1–26. [Google Scholar] [PubMed]
- Rahman, F.; Langford, K.H.; Scrimshaw, M.D.; Lester, J.N. Polybrominated diphenyl ether (PBDE) flame retardants. Sci. Total Environ. 2001, 275, 1–17. [Google Scholar] [CrossRef]
- Alan Wood’s Web Site. Compendium of Pesticide Common Names. Herbicides. Available online: http://www.alanwood.net/pesticides/class_herbicides.html#diphenyl_ether_herbicides (accessed on 31 January 2020).
- Kojima, H.; Iida, M.; Katsura, E.; Kanetoshi, A.; Hori, Y.; Kobayashi, K. Effects of a diphenyl ether-type herbicide, chlornitrofen, and its amino derivative on androgen and estrogen receptor activities. Environ. Health Perspect. 2003, 111, 497–502. [Google Scholar] [CrossRef] [Green Version]
- Keum, Y.S.; Lee, Y.J.; Kim, J.-H. Metabolism of nitrodiphenyl ether herbicides by dioxin-degrading bacterium Sphingomonas wittichii RW1. J. Agric. Food. Chem. 2008, 56, 9146–9151. [Google Scholar] [CrossRef]
- Im, J.; Löffer, F.E. Fate of bisphenol A in terrestrial and aquatic environments. Environ. Sci. Technol. 2016, 50, 8403–8416. [Google Scholar] [CrossRef]
- Lobos, J.H.; Leib, T.K.; Su, T.-M. Biodegradation of bisphenol A and other bisphenols by a gram-negative aerobic bacterium. Appl. Environ. Microbiol. 1992, 58, 1823–1831. [Google Scholar] [CrossRef] [Green Version]
- Spivack, J.; Leib, T.K.; Lobos, J.H. Novel pathway for bacterial metabolism of bisphenol A. J. Biol. Chem. 2005, 269, 7323–7329. [Google Scholar]
- Sasaki, M.; Akahira, A.; Oshima, K.; Tsuchido, T.; Matsumura, Y. Purification of cytochrome P-450 and ferredoxin, involved in bisphenol A degradation, from Sphingomonas sp. strain AO1. Appl. Environ. Microbiol. 2005, 71, 8024–8030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, J.; Kappelmeyer, U.; Kastner, M.; Schauer, F.; Heipieper, H.J. The degradation of bisphenol A by the newly isolated bacterium Cupriavidus basilensis JF1 can be enhanced by biostimulation with phenol. Int. Biodeterior. Biodegr. 2010, 64, 324–330. [Google Scholar] [CrossRef]
- Ogata, Y.; Goda, S.; Toyama, T.; Sei, K.; Ike, M. The 4-tert-butylphenol-utilizing bacterium Sphingobium fuliginis OMI can degrade bisphenols via phenolic ring hydroxylation and meta-cleavage pathway. Environ. Sci. Technol. 2013, 47, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Ike, M.; Chen, M.Y.; Danzl, E.; Sei, K.; Fujita, M. Biodegradation of a variety of bisphenols under aerobic and anaerobic conditions. Water Sci. Technol. 2006, 53, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danzl, E.; Sei, K.; Soda, S.; Ike, M.; Fujita, M. Biodegradation of bisphenol A, bisphenol F, and bisphenol S in seawater. Int. J. Environ. Res. Public Health 2009, 6, 1472–1484. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Yamanaka, H.; Moriyoshi, K.; Ohmoto, T.; Ohe, T. Biodegradation of bisphenol A and related compounds by Sphingomonas sp. strain BP-7 isolated from seawater. Biosci. Biotechnol. Biochem. 2007, 71, 51–57. [Google Scholar]
- Tanghe, T.; Dhooge, W.; Verstraete, W. Isolation of a bacterial strain able to degrade branched nonylphenol. Appl. Environ. Microbiol. 1999, 65, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, F.L.P.; Giger, W.; Guenther, K.; Kohler, H.E. Differential degradation of nonylphenol isomers by Sphingomonas xenophaga Bayram. Appl. Environ. Microbiol. 2005, 71, 1123–1129. [Google Scholar] [CrossRef] [Green Version]
- Poter, A.W.; Hay, A.G. Identification of opdA, a gene involved in biodegradation of the endocrine disrupter octylphenol. Appl. Environ. Microbiol. 2007, 73, 7373–7379. [Google Scholar] [CrossRef] [Green Version]
- Poter, A.W.; Campbell, B.R.; Kolvenbach, B.A.; Corvini, P.F.-X.; Benndorf, D.; Rivera-Cancel, G.; Hay, A.G. Identification of the flavin monooxygenase responsible for ipso substitution of alkyl and alkoxyphenols in Sphingomonas sp. TTNP3 and Sphingobium xenophagum Bayram. Appl. Microbiol. Biotechnol. 2012, 94, 261–272. [Google Scholar] [CrossRef]
- Ootsuka, M.; Nishizawa, T.; Hasegawa, M.; Kurusu, Y.; Ohta, H. Comparative analysis of the genetic basis of branched nonylphenol degradation by Sphingobium amiense DSM 16289T and Sphingobium cloacae JCM 10874T. Microbes. Environ. 2018, 33, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Liu, F.; Moon, H.-B.; Yamashita, N.; Yun, S.; Kannan, K. Bisphenol analogues in sediments from industrialized areas in the United States, Japan, and Korea: Spatial and temporal distributions. Environ. Sci. Technol. 2012, 46, 11558–11565. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, H.; Watanabe, M.; Shiraishi, F.; Shiraishi, H.; Shiozawa, T.; Matsushita, H.; Terao, Y. Formation of chlorinated derivatives of bisphenol A in waste paper recycling plants and their estrogenic activities. J. Health Sci. 2002, 48, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Yasahira, A.; Shiraishi, H.; Nakasugi, O. Bisphenol A in hazardous waste landfill leachates. Chemosphere 2001, 42, 415–418. [Google Scholar] [CrossRef]






| Compound | Initial Concentration (µM) 1 | Final Concentration (µM) 1 | Degradation Ratio (%) 2 | Detection of HQ |
|---|---|---|---|---|
| BPA | 200 ± 10 | 92 ± 6 | 54 | + |
| BPC | 258 ± 8 | 142 ± 7 | 45 | − |
| BPE | 139 ± 4 | 46 ± 2 | 67 | + |
| BPF | 139 ± 4 | 0 | 100 | + |
| BPS | 172 ± 8 | 40 ± 16 | 77 | + |
| BPZ | 193 ± 8 | 80 ± 6 | 59 | + |
| BP-AP | 273 ± 18 | 275 ± 5 | 0 | − |
| TP-PA | 141 ± 9 | 150 ± 18 | 0 | − |
| TDP | 182 ± 3 | 110 ± 2 | 40 | − |
| DBP | 143 ± 2 | 55 ± 2 | 62 | + |
| DDE | 179 ± 8 | 48 ± 2 | 73 | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeo, M.; Akizuki, J.; Kawasaki, A.; Negoro, S. Degradation Potential of the Nonylphenol Monooxygenase of Sphingomonas sp. NP5 for Bisphenols and Their Structural Analogs. Microorganisms 2020, 8, 284. https://doi.org/10.3390/microorganisms8020284
Takeo M, Akizuki J, Kawasaki A, Negoro S. Degradation Potential of the Nonylphenol Monooxygenase of Sphingomonas sp. NP5 for Bisphenols and Their Structural Analogs. Microorganisms. 2020; 8(2):284. https://doi.org/10.3390/microorganisms8020284
Chicago/Turabian StyleTakeo, Masahiro, Junichi Akizuki, Aika Kawasaki, and Seiji Negoro. 2020. "Degradation Potential of the Nonylphenol Monooxygenase of Sphingomonas sp. NP5 for Bisphenols and Their Structural Analogs" Microorganisms 8, no. 2: 284. https://doi.org/10.3390/microorganisms8020284
APA StyleTakeo, M., Akizuki, J., Kawasaki, A., & Negoro, S. (2020). Degradation Potential of the Nonylphenol Monooxygenase of Sphingomonas sp. NP5 for Bisphenols and Their Structural Analogs. Microorganisms, 8(2), 284. https://doi.org/10.3390/microorganisms8020284