Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Bacteria
2.3. PCR Confirmation of Species and Detection of AMR Genes
2.4. Antimicrobial Susceptibility Phenotyping
2.5. Whole-Genome Sequencing and Analyses
2.6. Statistical Analyses
3. Results
3.1. Bacterial Isolation, AMR Phenotyping and PCR Detection of AMR/ICE-Core Genes
3.2. Effects of Management, Year of Study and AMR Genes on MICs
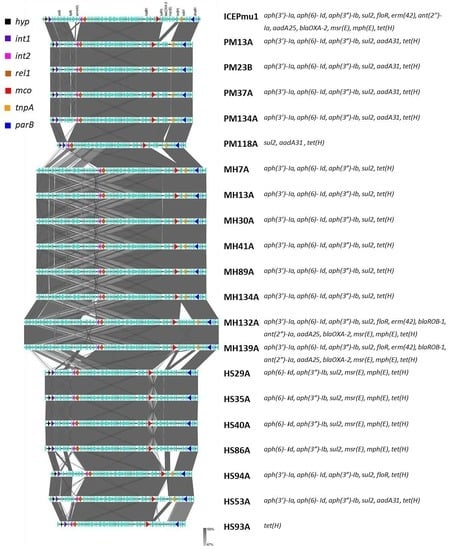
3.3. Sequence Analyses
4. Discussion
4.1. Recovery of Isolates
4.2. Year and Management Effects on PCR Detection of AMR and ICE Genes
4.3. Concordance of AMR Phenotype and Genotype
4.4. Factors Affecting MICs
5. Conclusions
Genome Sequence Data Availability
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crosby, S.; Credille, B.; Giguere, S.M.; Berghaus, R. Comparative efficacy of enrofloxacin to that of tulathromycin for the control of bovine respiratory disease and prevalence of antimicrobial resistance in Mannheimia haemolytica in calves at high risk of developing bovine respiratory disease. J. Anim. Sci. 2018, 96, 1259–1267. [Google Scholar] [CrossRef]
- Klima, C.L.; Zaheer, R.; Cook, S.R.; Booker, C.W.; Hendrick, S.; Alexander, T.W.; McAllister, T.A. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 2014, 52, 438–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, B.K.; Richards, C.J.; Step, D.L.; Krehbiel, C.R. Best management practices for newly weaned calved for improved health and well-being. J. Anim. Sci. 2017, 95, 2170–2182. [Google Scholar] [PubMed]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can. Vet. J. 2010, 51, 1095–1102. [Google Scholar]
- Griffin, D.; Chengappa, M.M.; Kuszak, J.; McVey, D.S. Bacterial pathogens of the bovine respiratory complex. Vet. Clin. Food Anim. 2010, 26, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Booker, C.W.; Abutarbush, S.M.; Morley, P.S.; Kee Jim, G.; Pittman, T.J.; Schunicht, O.C.; Perrett, T.; Wildman, B.K.; Fenton, R.K.; Guichon, P.T.; et al. Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in western Canada. Can. Vet. J. 2008, 49, 473–481. [Google Scholar] [PubMed]
- Fulton, R.W.; Blood, K.S.; Panciera, R.J.; Payton, M.E.; Ridpath, J.F.; Confer, A.W.; Saliki, J.T.; Burge, L.T.; Welsh, R.D.; Johnson, B.J.; et al. Lung pathology and infectious agents in fatal feedlot pneumonias and relationship with mortality, disease onset, and treatments. J. Vet. Diagn. Investig. 2009, 21, 464–477. [Google Scholar] [CrossRef] [Green Version]
- Welsh, R.D.; Dye, L.B.; Paton, M.E.; Confer, A.W. Isolation and antimicrobial susceptibility of bacterial pathogens from bovine pneumonia: 1994–2002. J. Vet. Diagn. Investig. 2004, 16, 426–431. [Google Scholar] [CrossRef] [Green Version]
- Gagea, M.I.; Bateman, K.G.; van Dreumel, T.; McEwen, B.J.; Archambault, M.; Shanahan, R.A.; Caswell, J.L. Diseases and pathogens associated with mortality in Ontario beef feedlots. J. Vet. Diagn. Investig. 2006, 18, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Portis, E.; Lindeman, C.; Johansen, L.; Stoltman, G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex–Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni–in the United States and Canada. J. Vet. Diagn. Investig. 2012, 24, 932–944. [Google Scholar] [CrossRef] [Green Version]
- Watts, J.L.; Sweeney, M.T. Antimicrobial resistance in bovine respiratory pathogens: Measures, trends, and impact on efficacy. Vet. Clin. Food Anim. 2010, 26, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Noyes, N.R.; Benedict, K.M.; Gow, S.P.; Booker, C.W.; Hannon, S.J.; McAllister, T.A.; Morley, P.S. Mannheimia haemolytica in feedlot cattle: Prevalence or recovery and associations with antimicrobial use, resistance and health outcomes. J. Vet. Intern. Med. 2015, 29, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Ayling, R.D.; Rosales, R.S.; Barden, G.; Gosney, F.L. Changes in antimicrobial susceptibility of Mycoplasma bovis isolates from Great Britain. Vet. Record 2014, 175, 486. [Google Scholar] [CrossRef] [PubMed]
- DeDonder, K.D.; Apley, M.D. A literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease. Anim. Health Res. Rev. 2015, 16, 125–134. [Google Scholar] [CrossRef]
- Anholt, R.M.; Klima, C.; Allan, N.; Matheson-Bird, H.; Schatz, C.; Ajilkumar, P.; Otto, S.J.G.; Peters, D.; Schmid, K.; Olson, M.; et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta Canada. Front. Vet. Sci. 2017, 4, 207. [Google Scholar] [CrossRef] [Green Version]
- Federation of Veterinarians of Europe. Antimicrobial Use in Food-Producing Animals. Replies to EFSA/EMA Questions on the Use of Antimicrobials in Food-Producing Animals in EU and Possible Measures to Reduce Antimicrobial Use; Federation of Veterinarians of Europe: Brussels, Belgium, 2016; p. 91. [Google Scholar]
- Bergman, M.; Nyberg, S.T.; Huovinen, P.; Paakkari, P.; Hakanen, A.J. Association between antimicrobial consumption and resistance in Escherichia coli. Antimicrob. Agents Chemother. 2009, 53, 912–917. [Google Scholar] [CrossRef] [Green Version]
- Ismael, A.B.; Hassan, M.Y.; Mostafa, S.A.-H.; Nassan, M.A.; Mohamed, E.H. Development of a second-generation vaccine against Mycoplasmosis: Preparation of a fraction candidate from Mycoplasma bovis and its evaluation as a vaccine. Global Vet. 2016, 16, 137–144. [Google Scholar]
- Gioia, G.; Werner, B.; Nydam, D.V.; Moroni, P. Validation of a mycoplasma molecular diagnostic test and distribution of mycoplasma species in bovine milk among New York state dairy farms. J. Dairy Sci. 2016, 99, 4668–4677. [Google Scholar] [CrossRef]
- Michael, G.B.; Kadlec, K.; Sweeney, M.T.; Bzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Murray, R.W.; Watts, J.L.; Schwartz, S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Structure and transfer. J. Antimicrob. Chemother. 2012, 67, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistance genes. Mol. Cell. Probes 2001, 15, 209–2215. [Google Scholar] [CrossRef]
- Klima, C.L.; Alexander, T.W.; Read, R.R.; Gow, S.P.; Booker, C.W.; Hannon, S.; Sheedy, C.; McAllister, T.A.; Selinger, L.B. Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet. Microbiol. 2011, 149, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Klima, C.L.; Zaheer, R.; Briggs, R.E.; McAllister, T.A. A multiplex PCR assay for molecular capsular serotyping of Mannheimia haemolytica serotypes 1, 2, and 6. J. Microbiol. Methods 2017, 139, 155–160. [Google Scholar] [CrossRef]
- Kolaj-Robin, O.; Russell, D.; Hayes, K.A.; Pembroke, J.T.; Soulimane, T. Cation diffusion facilitator family: Structure and function. FEBS Lett. 2015, 589, 1283–1295. [Google Scholar] [CrossRef]
- Beker, M.; Rose, S.; Lykkebo, C.A.; Douthwaite, S. Integrative and conjugative elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front. Microbiol. 2018, 9, 1329. [Google Scholar] [CrossRef]
- Stothard, P.; Grant, J.R.; van Domselaar, G. Visualizing and comparing circular genomes using CGView family of tools. Brief. Bioinform. 2019, 20, 1576–1582. [Google Scholar] [CrossRef] [Green Version]
- Snowder, G.D.; Van Vleck, L.D.; Cundiff, L.V.; Bennet, G.L. Bovine respiratory disease in feedlot cattle: Environmental, genetic and economic factors. J. Anim. Sci. 2006, 84, 1999–2008. [Google Scholar] [CrossRef] [Green Version]
- Timsit, E.; Christensen, H.; Bareille, N.; Seegers, H.; Bisgaard, M.; Assie, S. Transmission dynamics of Mannheimia haemolytica in newly-received beef bulls at fattening operations. Vet. Microbiol. 2013, 161, 295–304. [Google Scholar] [CrossRef]
- Stroebel, C.; Alexander, T.; Workentine, M.; Timsit, E. Effects of transportation to and co-mingling at an auction market on nasopharyngeal and tracheal bacterial communities of recently weaned beef cattle. Vet. Microbiol. 2018, 223, 126–133. [Google Scholar] [CrossRef]
- Timsit, E.; Workentine, M.; Crepiuex, T.; Miller, C.; Regev-Shoshani, G.; Schaefer, A.; Alexander, T. Evolution of the nasopharangeal microbiota of beef cattle from weaning to 40 days after arrival at a feedlot. Vet. Microbiol. 2016, 187, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; McAllister, T.A.; Topp, E.; Wright, A.D.G.; Alexander, T.W. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet. Microbiol. 2015, 180, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Klima, C.L.; Holman, D.B.; Ralston, B.J.; Stanford, K.; Zaheer, R.; Alexander, T.W.; McAllister, T.A. Lower respiratory tract microbiome and resistome of bovine respiratory disease mortalities. Microbiol. Ecol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cusack, P.M.V.; McMeniman, N.; Lean, I.J. The medicine and epidemiology of bovine respiratory disease in feedlots. Aust. Vet. J. 2003, 81, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Michael, G.B.; Kadlec, K.; Sweeney, M.T.; Brzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Murray, R.W.; Watts, J.L.; Schwartz, S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.R.; Noyes, N.; Young, A.E.; Prince, D.J.; Blanchard, P.C.; Lehenbauer, T.W.; Aly, S.S.; Davis, H.H.; O’Rourke, S.M.; Abdo, Z.; et al. Whole genome sequencing and concordance between antimicrobial susceptibility genotypes and phenotypes of bacterial isolates associated with bovine respiratory disease. G3 2017, 7, 3059–3071. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, K.; Timsit, E.; Rawlyk, N.; Potter, A.; Lifebjelke, K. Integrative conjugative element ICEHs1 encodes for antimicrobial resistance and metal tolerance in Histophilus somni. Front. Vet. Sci. 2018, 5, 153. [Google Scholar] [CrossRef]
- Lysnyansky, I.; Ayling, R.D. Mycoplasma bovis: Mechanisms of resistance and trends in antimicrobial susceptibility. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Sulyok, K.M.; Kreizinger, Z.; Whmann, E.; Lysnyansky, O.; Banyai, K.; Marton, S.; Jerzsele, A.; Ronai, Z.; Turcsanyi, I.; Makrai, L.; et al. Mutations associated with decreased susceptibility to seven antimicrobial families in field and laboratory-derived Mycoplasma bovis strains. Antimicrob. Agents Chemo. 2017, 61, e01983-16. [Google Scholar] [CrossRef] [Green Version]
- Calcutt, M.J.; Lysnyansky, I.; Sachse, K.; Fox, L.K.; Nicholas, R.A.J.; Ayling, R.D. Gap analysis of Mycoplasma bovis disease, diagnosis and control: An aid to identify future development requirements. Transbound. Emerg. Dis. 2018, 65 (Suppl. 1), 91–109. [Google Scholar] [CrossRef] [Green Version]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Dettenwanger, A.; Cook, S.; Geornaras, I.; Woerner, D.E.; Gow, S.P.; McAllister, T.A.; et al. Resistome diversity in cattle and the environment decreases during beef production. eLife 2016, 5, e13195. [Google Scholar] [CrossRef]
- Morley, P.S.; Dargatz, D.A.; Hyatt, D.R.; Dewell, G.A.; Patterson, J.G.; Burgess, B.A.; Wittum, T.E. Effects of restricted antimicrobial exposure on antimicrobial resistance in fecal Escherichia coli from feedlot cattle. Foodborne Path. Dis. 2011, 8, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Wall, S.K.; Xu, L.; Ebner, P.D. Contamination rates and antimicrobial resistance in bacteria isolated from “grass-fed” labeled beef products. Foodborne Path. Dis. 2010, 7, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Schmidt, J.W. Functional blaKPC-2 sequences are present in U.S. beef cattle feces regardless of antibiotic use. Foodborne Path. Dis. 2018, 15, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Rovira, P.; Agga, G.E.; Arthur, T.M.; Bosilevac, J.M.; Wheeler, T.L.; Morley, P.S.; Belk, K.E.; Schmidt, J.W. Impact of “Raised without antibiotics” beef cattle production practices on occurrences of antimicrobial resistance. Appl. Environ. Microbiol. 2017, 83, e01682-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klima, C.L.; Cook, S.R.; Zaheer, R.; Laing, C.; Gannon, V.P.; Xu, Y.; Rasmussen, J.; Potter, A.; Hendrick, S.; Alexander, T.W.; et al. Comparative genomic analysis of Mannheimia haemolytica from bovine sources. PLoS ONE 2016, 11, e0149520. [Google Scholar] [CrossRef] [PubMed]
- Clawson, M.L.; Murray, R.W.; Sweeney, M.T.; Apley, M.D.; DeDonder, K.D.; Capik, S.F.; Larson, R.L.; Lubbers, B.V.; White, B.J.; Kalbfleisch, T.S.; et al. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics. 2016, 17, 982. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.; Klima, C.L.; Ha, R.; Gruninger, R.J.; Zaheer, R.; McAllister, T.A. A novel aadA aminoglycoside resistance gene in bovine and porcine pathogens. mSphere 2018, 3, e00568-17. [Google Scholar] [CrossRef] [Green Version]
- Kehrenberg, C.; Schwarz, S. Mutations in 16S rRNA and ribosomal protein S5 associated with high-level spectinomycin resistance in Pasteurella multocida. Antimicrob. Agents Chemother. 2007, 51, 2244–2246. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.S.; Warrass, R.; Douthwaite, S. Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 2015, 70, 420–423. [Google Scholar] [CrossRef]
- Vester, B.; Douthwaite, S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001, 45, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.C.; Gao, D.; Gao, Y.H.; Liu, S.M.; Ma, H.X. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J. Vet. Med. Sci. 2014, 76, 1655–1657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozawa, M.; Asai, T.; Sameshima, T. Mutations in GyrA and ParC in fluoroquinolone-resistant Mannheimia haemolytica isolates from cattle in Japan. J. Vet. Med. Sci. 2009, 71, 493–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkey, P.M. Mechanisms of quinolone action and microbial response. J. Antimicrob. Chemother. 2003, 51 (Suppl. 1), 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Martínez, J.M.; Cano, M.E.; Velasco, C.; Martínez-Martínez, L.; Pascual, A. Plasmid-mediated quinolone resistance: An update. J. Infect. Chemother. 2011, 17, 149–182. [Google Scholar] [CrossRef] [PubMed]
- Seyedpour, S.M.; Eftekhar, F. Quinolone susceptibility and detection of qnr and aac(6′)-Ib-cr genes in community isolates of Klebsiella pneumoniae. Jundishapur J. Microbiol. 2014, 7, e11136. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Martínez, L.; Eliecer Cano, M.; Rodríguez-Martínez, M.J.; Calvo, J.; Pascual, A. Plasmid-mediated quinolone resistance. Expert Rev. Anti-Infect Ther. 2008, 6, 685–711. [Google Scholar] [CrossRef]
- Piekarska, K.; Wołkowicz, T.; Zacharczuk, K.; Rzeczkowska, M.; Chróst, A. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int. J. Antimicrob. Agents 2015, 45, 238–243. [Google Scholar] [CrossRef]
- Gautier-Bouchardon, A.V.; Ferre, S.; Le Grand, D.; Paoli, A.; Gay, E.; Pourmarat, F. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE 2014, 9, e87672. [Google Scholar] [CrossRef] [Green Version]
- Doster, E.; Rovira, P.; Noyes, N.R.; Burgess, B.A.; Yang, X.; Weinroth, M.D.; Lakin, S.M.; Dean, C.J.; Linke, L.; Magnuson, R.; et al. Investigating effects of tulathromycin metaphylaxis on the fecal resistome and microbiome of commercial cattle early in the feeding period. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Rajamanickam, K.; Yang, J.; Sakharkar, M.S. Gallic acid potentiates the antimicrobial activity of tulathromycin against two key bovine respiratory disease (BRD) causing pathogens. Front. Pharma. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Maunsell, F.P.; Woolums, A.R.; Francoz, D.; Rosenbusch, R.F.; Step, D.L.; Wilson, D.J.; Janzen, E.D. Mycoplasma bovis infections in cattle. J. Vet. Intern. Med. 2011, 25, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Health Canada. Uses of Antimicrobials in Food Animals in Canada: Impact on Resistance and Human Health. 2002. Available online: www.canada.ca/en/health-canada (accessed on 12 November 2019).
- Demoliaze, B.; Rose, S.; Warrass, R.; Douthwaite, S. A novel Erm monomethyltransferase in antibiotic-resistant isolates of Mannheimia haemolytica and Pasteurella multocida. Mol. Microbiol. 2011, 80, 184–194. [Google Scholar]
- Eidam, C.; Poehlein, A.; Leimbach, A.; Michael, G.B.; Kadlec, K.; Liesegang, H.; Daniel, R.; Sweeney, M.T.; Murray, R.W.; Watts, J.L.; et al. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 2015, 70, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Phenotype/Function | PCR Target(s) | Base Pairs | T z | Reference |
|---|---|---|---|---|
| Antimicrobial resistance class | ||||
| Aminoglycoside | ||||
| gentamicin | ant(2″)-Ia | 551 | 66 | [2] |
| neomycin | aph(3′)-Ia | 489 | 54 | [21] |
| spectinomycin | aadA25 | 503 | 66 | [2] |
| streptomycin | aph(3″)-Ib | 506 | 64 | [2] |
| aph(6)-Id | 586 | 64 | [2] | |
| Amphenicol | floR | 320 | 58 | [2] |
| Macrolide | msr(E) mph(E) erm(42) | 620,401, 1254 | 60 | [2] |
| Penicillin | blaOXA-2 | 685 | 60 | [22] |
| Sulfonamide | sul2 | 489 | 64 | [2] |
| Tetracycline | tet(H) tet(R) | 1076 | 60 | [22] |
| Ice core genes | ||||
| Hypothetical protein | Pmu02680 | 226 | 58 | [2] |
| Integrase (int1) | Pmu02700 | 735 | 58 | [2] |
| Multi-copper oxidase (mco) | Pmu03360 | 458 | 58 | [2] |
| Transposase (tnpA) | Pmu03510 | 204 | 56 | [2] |
| ICE-Relaxase (rel) | Pmu02890 | 695 | 57 | [20] |
| Organism | # of Isolates Collected | Overall Prevalence (%) | # of Isolates with Sensititre Analyses z | # NAT Isolates with Sensititre Analyses |
|---|---|---|---|---|
| Mannheimia haemolytica | 113 | 31.6 ± 2.2 b | 104 | 6 |
| Pasteurella multocida | 47 | 12.9 ± 2.2 a | 45 | 4 |
| Histophilus somni | 41 | 12.5 ± 1.4 a | 23 y | 0 |
| Mycoplasma bovis | 227 | 63.9 ± 3.8 c | 61 x | 7 |
| Organism | Management z | Year of Study | SEM | ||
|---|---|---|---|---|---|
| CON | NAT | 1 | 2 | ||
| Mannheimia haemolytica | 6.7 | 4.1 | 2.6 a | 8.2 b | 2.8 |
| Pasteurella multocida | 4.4 b | 0.5 a | 3.9 | 3.8 | 2.3 |
| Histophilus somni | 2.8 | NA y | 2.8 | NA | NA |
| Strain | hyp | int | rlx | mco | tnp | aph(3′) Ia | aph(6)Id | aph(3″)Ib | sul2 | floR | erm (42) | ant(2″)Ia | aadA25 | bla OXA-2 | msr (E) | mph (E) | tet (R) | tet (H) | Total Genes | # Isolates with Same Pattern (% of Total Isolates) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MH | - | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 16 | 44/111 (39.6) |
| MH | - | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1 | 23/111 (20.7) |
| MH | - | - | + | + | + | + | + | + | + | - | - | - | - | - | - | - | + | - | 8 | 14/111 (12.6) |
| PM | + | + | + | + | + | + | + | + | + | - | - | - | - | - | - | - | + | + | 11 | 22/47 (46.8) |
| PM | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0 | 10/47(21.3) |
| PM | + | + | + | + | + | - | - | - | - | - | - | - | - | - | - | - | + | + | 7 | 9/47(19.1) |
| HS | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0 | 11/42(26.2) |
| HS | + | + | + | + | - | - | - | - | + | + | - | - | - | - | + | + | + | + | 10 | 7/42 (16.7) |
| HS | + | + | + | + | - | + | + | + | + | + | - | - | - | - | + | + | + | + | 13 | 3/42 (7.1) |
| Antimicrobial | CON | NAT | 1 | 2 | SEM | Gene | + | - | Gene | + | - | Gene | + | - |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | 15.4 | 14.3 | 14.1 | 15.7 | NA y | blaOX-A2 | 29.5 | 0.2 | relaxase | 14.8 | 14.9 | mco | 15.2 | 14.1 |
| Ceftiofur | 0.2 | 0.0 | 0.4 | 0.0 | 0.9 | relaxase | 0.6 | 0.0 | mcox | 0.0 | 0.2 | |||
| Clindamycin | 20.3 | 16.2 | 16.5 a | 19.9 b | 2.3 | relaxase | 27.3 b | 9.2 a | mco | 18.2 | 18.2 | |||
| Chlortetracycline | 2.1 | 1.9 | 1.5 a | 2.7 b | 0.6 | tet(H) | 2.2 | 1.9 | tet(R) | 1.7 | 2.4 | relaxase | 2.5 | 1.6 |
| mco | 4.4 b | 0.9 a | ||||||||||||
| Danofloxacin | 0.6 | 0.3 | 0.1 | 0.8 | NA | relaxase | 0.4 | 0.5 | mco | 0.9 | 0.0 | |||
| Enrofloxacin | 1.2 | 0.7 | 0.2 | 1.6 | NA | relaxase | 1.1 | 0.7 | mco | 1.9 | 0.0 | |||
| Florfenicol | 3.7 | 3.8 | 4.1 | 3.4 | 0.6 | floR | 14.6 b | 0.9 a | relaxase | 4.4 | 3.1 | mco | 3.5 | 4.0 |
| Gentamicin | 11.2 | 11.8 | 11.6 | 11.4 | 2.2 | ant(2″)-Ia | 43.7 b | 3.0 a | relaxase | 15.1 b | 8.8 a | mco | 11.2 | 11.9 |
| Neomycin | 23.0 | 22.3 | 22.6 | 22.7 | 0.8 | aph(3″)Ia | 25.2 b | 20.3 a | relaxase | 22.7 | 22.6 | mco | 63.3 b | 8.1 a |
| Oxytetracycline | 3.5 | 3.3 | 1.7 a | 4.2 b | 0.9 | tet(H) | 4.1 | 2.8 | tet(R) | 3.0 | 3.8 | relaxase | 4.4 | 2.6 |
| mco | 10.2 b | 1.1 a | ||||||||||||
| Penicillin | 7.5 | 7.0 | 6.9 | 7.7 | NA | blaOX-A2 | 13.4 | 1.2 | relaxase | 7.2 | 7.3 | mco | 7.5 | 7.0 |
| Spectinomycin | 77.6 | 50.7 | 63.1 | 65.3 | 11.4 | aadA25 | 98.0 b | 30.3 a | relaxase | 61.2 | 67.1 | mco | 72.9 b | 55.4 a |
| Sullfadimethoxine | 254.6 | 206.4 | 228.8 | 232.2 | NA | sul2 | 227.9 | 233.0 | relaxase | 228.6 | 232.5 | mco | 233.1 | 228.0 |
| Tilmicosin | 63.2 b | 30.2 a | 48.2 | 45.2 | 12.5 | msr(E) | 89.6 b | 3.8 a | mphE | 55.7 | 37.7 | erm(42) | 76.5 b | 17.0 a |
| relaxase | 52.2 | 41.2 | mco | 84.3 b | 9.1 a | |||||||||
| Trimethroprim/ Sulfmethazole | 0.05 | 0.02 | 0.04 | 0.03 | NA | sul2 | 0.02 | 0.05 | relaxase | 0.07 | 0.00 | mco | 0.06 | 0.02 |
| Tulathromycin | 61.1 b | 28.9 a | 45.8 | 28.9 | 12.8 | msr(E) | 86.1 b | 3.9 a | mph(E) | 53.1 | 36.9 | erm(42) | 72.7 b | 17.4 a |
| relaxase | 54.8 | 35.2 | mco | 54.8 b | 9.3 a | |||||||||
| Tylosin | 63.0 | 67.0 | 60.9 | 69.1 | NA | msr(E) | 66.2 | 63.8 | mph(E) | 63.2 | 66.2 | erm(42) | 55.1 | 65.2 |
| relaxase | 62.8 | 67.2 | mco | 68.4 | 62.8 |
| Antimicrobial | CON | NAT | 1 | 2 | SEM | Gene | + | - | Gene | + | - | Gene | + | - |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | 11.7 | 21.3 | 10.7 a | 22.3 b | 6.1 | blaOXA-2 | NAv | ICE | 20.0 | 13.0 | ||||
| Ceftiofur | 6.1 | 11.1 | 7.7 | 9.4 | 3.2 | ICEy | 11.0 b | 6.1 a | ||||||
| Chlortetracycline | 2.1 | 0.4 | 1.3 | 0.7 | 1.5 | tet(H), tet(R) | 0.8 | 1.1 | ICE | NAx | ||||
| Clindamycin | 32.0 | 32.0 | 32.0 | 32.0 | NAw | ICE | 32.0 | 32.0 | ||||||
| Danofloxacin | 0.33 | 0.20 | 0.24 | 0.29 | 0.29 | ICE | 0.44 | 0.09 | ||||||
| Enrofloxacin | 0.13 | 0.06 | 0.12 | 0.07 | 0.39 | ICE | 0.17 | 0.02 | ||||||
| Florfenicol | 1.0 | 1.2 | 1.4 | 0.8 | 1.2 | floR | NAu | ICE | 1.4 | 0.9 | ||||
| Gentamicin | 5.4 a | 17.1 b | 10.7 | 11.9 | 3.3 | ant(2″)-Ia | NAw | ICE | 11.8 | 10.8 | ||||
| Neomycin | 43.2 b | 20.9 a | 34.3 | 29.1 | 6.1 | aph(3′)Ia | 55.9 a | 8.1 b | ICE | 27.3 | 36.8 | |||
| Oxytetracycline | 8.1 | 7.6 | 9.1 | 6.5 | 3.7 | tet(H), tet(R) | 10.2 | 5.4 | ICE | NAx | ||||
| Penicillin | 5.3 | 7.7 | 4.9 | 8.1 | 3.8 | blaOXA-2 | NAv | ICE | 8.3 | 4.7 | ||||
| Spectinomycin | 94.0 a | 37.8 b | 69.0 | 57.9 | 11.2 | aadA25 | NAu | ICE | 93.3 b | 33.5 a | ||||
| Sulfadimethoxine | 256 | 256 | 256 | 256 | NAw | sul2 | 256 | 256 | ICE | 256 | 256 | |||
| Tilmicosin | 66.6 | 54.4 | 70.4 | 50.6 | 25.8 | msr(E) mph(E) | NAu | erm(42) | NAu | ICE | 94.2 b | 26.8 a | ||
| Trimethroprim/ Sulfmethazole | 0.08 | 0.09 | 0.09 | 0.08 | NAw | sul2 | 0.09 | 0.08 | ICE | 0.10 | 0.07 | |||
| Tulathromycin | 63.3 | 61.5 | 72.0 | 52.8 | 26.1 | msr(E) mph(E) | NAu | erm(42) | NAu | ICE | 99.6 b | 25.2 a | ||
| Tylosin | 48.3 | 41.1 | 48.2 | 41.2 | NAw | msr(E) mph(E) | NAu | erm(42) | NAu | ICE | 57.2 | 32.1 | ||
| Antimicrobial | Gene | + | - | Gene | + | - | Gene | + | - | Gene | + | - |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | blaOXA-2 | NAx | ICEz | 4.0 | 0.0 | TNP | 0.0 | 3.4 | ||||
| Ceftiofur | ICE | 0.08 | 0.03 | TNP | 0.11 | 0.00 | ||||||
| Chlortetracycline | tet(H) | 1.8 | 0.9 | tet(R)/ICEy | 3.0 b | 0.0 a | TNP | 2.3 b | 0.3 a | |||
| Clindamycin | ICE | 1.3 | 0.8 | TNP | 0.9 | 1.2 | ||||||
| Danofloxacin | ICE | 0.19 | 0.00 | TNP | 0.0 | 0.14 | ||||||
| Enrofloxacin | ICE | 0.27 | 0.00 | TNP | 0.0 | 0.22 | ||||||
| Florfenicol | floR | 4.4 b | 0.3 a | ICE | 2.2 | 2.5 | TNP | 2.6 | 2.1 | |||
| Gentamicin | ant(2″)-Ia | NAw | ICE | 17.2 | 10.3 | TNP | 13.3 | 14.2 | ||||
| Neomycin | aph(3′)-Ia | 42.8 | 40.3 | ICE | 51.2 | 31.9 | TNP | 34.9 | 48.2 | |||
| Oxytetracycline | tet(H) | 6.0 | 6.7 | tet(R)/ICE | 13.9 | 0.0 | TNP | 4.6 | 8.1 | |||
| Penicillin | blaOXA-2 | NAx | ICE | 5.4 | 0.0 | TNP | 2.0 | 2.6 | ||||
| Spectinomycin | aadA25 | NAx | ICE | 35.4 | 5.6 | TNP | 19.7 | 15.2 | ||||
| Sullfadimethoxine | sul2 | 222.8 | 217.6 | ICE | 252.2 | 188.2 | TNP | 207.3 | 233.1 | |||
| Tilmicosin | msr(E) or mph(E) | 78.6 b | 3.0 a | erm(42) | 43.1 | 38.6 | ICE | 51.2 | 31.9 | TNP | 10.7 | 11.4 |
| Trimethroprim/ Sulfmethazole | sul2 | 0.01 | 0.00 | ICE | 0.01 | 0.00 | TNP | 0.00 | 0.01 | |||
| Tulathromycin | msr(E) or mph(E) | 127.5 | 5.8 | erm(42) | 68.0 | 65.2 | ICE | 68.1 | 65.1 | TNP | 66.0 | 67.3 |
| Tylosin | msr(E) or mph(E) | 15.52 | 4.7 | erm(42) | 8.4 | 11.8 | ICE | 18.2 b | 2.0 a | TNP | 7.4 | 12.8 |
| Antimicrobial | CON | NAT | 1 | 2 | SEM |
|---|---|---|---|---|---|
| Chlortetracycline | 4.2 | 3.3 | 3.8 | 3.6 | 0.9 |
| Enrofloxacin | 1.3 | 0.2 | 0.7 | 0.8 | 1.1 |
| Florfenicol | 4.4 | 2.9 | 3.7 | 3.6 | 0.9 |
| Gamithromycin | 239.0 b | 162.4 a | 245.9 b | 155.6 a | 12.3 |
| Oxytetracycline | 4.1 | 3.2 | 3.2 | 4.2 | 0.7 |
| Tilmicosin | 253.3 | 256.0 | 253.3 | 256.0 | NA |
| Tidipirosin | 251.1 | 256.0 | 251.1 | 256.0 | NA |
| Tulathromycin | 225.9 b | 141.3 a | 239.2 b | 128.0 a | 20.8 |
| Tylosin | 198.7 | 183.5 | 236.9 | 145.3 | 24.8 |
| Strain | aph (3′)-Ia | aph (6)-Id | aph (3″)-Ib | sul2 | floR | erm (42) | ant (2″)-Ia | aad A31 | aad A25 | blaOXA-2 | blaROB-1 | mph (E) | msr (E) | tet (H) | QRDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MH-1 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-2 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-3 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-4 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-5 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-6 | + | + | + | + | - | - | - | - | - | - | - | - | - | + | - |
| MH-7 | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + (GyrAy: S83F, D87N; ParCy: E89K) |
| MH-8 | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + (GyrA: S83F, D87N; ParC: E89K) |
| MH-9 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PM-1 | + | + | + | + | - | - | - | + | - | - | - | - | - | + | - |
| PM-2 | + | + | + | + | - | - | - | + | - | - | - | - | - | + | - |
| PM-3 | + | + | + | + | - | - | - | + | - | - | - | - | - | + | - |
| PM-4 | - | - | - | - | - | - | - | + | - | - | - | - | - | + | (GyrA: S88R) |
| PM-5 | + | + | + | + | - | - | - | + | - | - | - | - | - | + | - |
| HS-1 | - | + | + | + | - | - | - | - | - | - | - | + | + | + | - |
| HS-2 | - | + | + | + | - | - | - | - | - | - | - | + | + | + | - |
| HS-3 | - | + | + | + | - | - | - | - | - | - | - | + | + | + | - |
| HS-4 | + | + | + | + | - | - | - | + | - | - | - | - | - | + | - |
| HS-5 | - | + | + | + | - | - | - | - | - | - | - | + | + | + | - |
| HS-6 | - | - | - | - | - | - | - | - | - | - | - | - | - | + | - |
| HS-7 | + | + | + | + | + | - | - | - | - | - | - | - | - | + | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanford, K.; Zaheer, R.; Klima, C.; McAllister, T.; Peters, D.; Niu, Y.D.; Ralston, B. Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials. Microorganisms 2020, 8, 288. https://doi.org/10.3390/microorganisms8020288
Stanford K, Zaheer R, Klima C, McAllister T, Peters D, Niu YD, Ralston B. Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials. Microorganisms. 2020; 8(2):288. https://doi.org/10.3390/microorganisms8020288
Chicago/Turabian StyleStanford, Kim, Rahat Zaheer, Cassidy Klima, Tim McAllister, Delores Peters, Yan D. Niu, and Brenda Ralston. 2020. "Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials" Microorganisms 8, no. 2: 288. https://doi.org/10.3390/microorganisms8020288
APA StyleStanford, K., Zaheer, R., Klima, C., McAllister, T., Peters, D., Niu, Y. D., & Ralston, B. (2020). Antimicrobial Resistance in Members of the Bacterial Bovine Respiratory Disease Complex Isolated from Lung Tissue of Cattle Mortalities Managed with or without the Use of Antimicrobials. Microorganisms, 8(2), 288. https://doi.org/10.3390/microorganisms8020288