In vitro Edwardsiella piscicida CK108 Transcriptome Profiles with Subinhibitory Concentrations of Phenol and Formalin Reveal New Insights into Bacterial Pathogenesis Mechanisms
Abstract
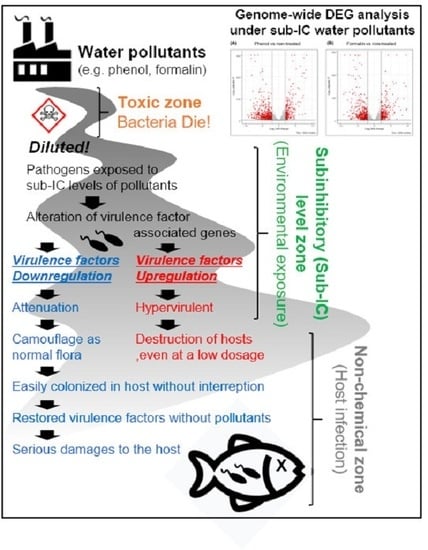
:1. Introduction
2. Materials and Methods
2.1. Strain and Growth Conditions
2.2. RNA Extraction, Library Construction, and Sequencing
2.3. RNA-seq Data Analysis
2.4. Quantitative Reverse Transcription PCR
2.5. Motility Assay
2.6. Data Availability
3. Results
3.1. Determination and Growth of E. piscicida CK108 in Sub-ICs of Phenol and Formalin
3.2. Overall Genome-Wide Gene Expression Profiling
3.3. Functional Enrichment Analysis with DEGs
3.4. Pathway Analysis
3.5. Validation of RNA-seq Data by qRT-PCR
3.6. Motility Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tisler, T.; Krel, A.; Gerzelj, U.; Erjavec, B.; Dolenc, M.S.; Pintar, A. Hazard identification and risk characterization of bisphenols A, F and AF to aquatic organisms. Environ. Pollut. 2016, 212, 472–479. [Google Scholar] [CrossRef]
- Sharifan, H. Alarming the impacts of the organic and inorganic UV blockers on endangered coral’s species in the Persian Gulf: A scientific concern for coral protection. Sustain. Futures 2020, 2, 100017. [Google Scholar] [CrossRef]
- Sharifan, H.; Ma, X.M. Potential Photochemical Interactions of UV Filter Molecules with Multichlorinated Structure of Prymnesins in Harmful Algal Bloom Events. Mini Rev. Org. Chem. 2017, 14, 391–399. [Google Scholar] [CrossRef]
- Li, H.; Meng, F.; Duan, W.; Lin, Y.; Zheng, Y. Biodegradation of phenol in saline or hypersaline environments by bacteria: A review. Ecotoxicol. Environ. Saf. 2019, 184, 109658. [Google Scholar] [CrossRef]
- Hinteregger, C.; Leitner, R.; Loidl, M.; Ferschl, A.; Streichsbier, F. Degradation of phenol and phenolic compounds by Pseudomonas putida EKII. Appl. Microbiol. Biotechnol. 1992, 37, 252–259. [Google Scholar] [CrossRef]
- Bruce, R.M.; Santodonato, J.; Neal, M.W. Summary review of the health effects associated with phenol. Toxicol. Ind. Health 1987, 3, 535–568. [Google Scholar] [CrossRef]
- Lauchnor, E.G.; Radniecki, T.S.; Semprini, L. Inhibition and gene expression of Nitrosomonas europaea biofilms exposed to phenol and toluene. Biotechnol. Bioeng. 2011, 108, 750–757. [Google Scholar] [CrossRef]
- Duan, W.; Meng, F.; Cui, H.; Lin, Y.; Wang, G.; Wu, J. Ecotoxicity of phenol and cresols to aquatic organisms: A review. Ecotoxicol. Environ. Saf. 2018, 157, 441–456. [Google Scholar] [CrossRef]
- Leal, J.F.; Neves, M.G.P.M.S.; Santos, E.B.H.; Esteves, V.I. Use of formalin in intensive aquaculture: Properties, application and effects on fish and water quality. Rev. Aquacult. 2018, 10, 281–295. [Google Scholar] [CrossRef]
- Park, I.S.; Baek, S.W.; Moon, K.H. The Sterilization Effect of Methylene Blue, Formalin, and Iodine on Egg and Adult Stage of Marine Medaka, Oryzias dancena. Dev. Reprod. 2019, 23, 199–211. [Google Scholar] [CrossRef]
- Park, S.B.; Aoki, T.; Jung, T.S. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 2012, 43, 67. [Google Scholar] [CrossRef] [Green Version]
- Bujan, N.; Toranzo, A.E.; Magarinos, B. Edwardsiella piscicida: A significant bacterial pathogen of cultured fish. Dis. Aquat. Organ. 2018, 131, 59–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, K.Y.; Wang, Q.; Yang, Z.; Siame, B.A. Edwardsiella piscicida: A versatile emerging pathogen of fish. Virulence 2019, 10, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, K.Y.; Siame, B.A.; Tenkink, B.J.; Noort, R.J.; Mok, Y.K. Edwardsiella tarda—Virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012, 14, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Alwine, J.C.; Kemp, D.J.; Stark, G.R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Nat. Acad. Sci. USA 1977, 74, 5350–5354. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.M.; Doyle, M.V.; Mark, D.F. Quantitation of mRNA by the polymerase chain reaction. Proc. Nat. Acad. Sci. USA 1989, 86, 9717–9721. [Google Scholar] [CrossRef] [Green Version]
- Schulze, A.; Downward, J. Navigating gene expression using microarrays—A technology review. Nat. Cell Biol. 2001, 3, E190–E195. [Google Scholar] [CrossRef]
- Hwang, S.; Chavarria, N.E.; Hackley, R.K.; Schmid, A.K.; Maupin-Furlow, J.A. Gene Expression of Haloferax volcanii on Intermediate and Abundant Sources of Fixed Nitrogen. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Yu, J.E.; Cho, M.Y.; Kim, J.W.; Kang, H.Y. Large antibiotic-resistance plasmid of Edwardsiella tarda contributes to virulence in fish. Microb. Pathog. 2012, 52, 259–266. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 12 June 2020).
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G., Jr.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef]
- Baek, S.W.; Hwang, S.; Kang, H.Y.; Bang, W.Y.; Moon, K.H. Draft Genome Sequence of a Fish Pathogen, Edwardsiella piscicida Isolate CK41. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Li, H.; Sun, B.; Ning, X.; Jiang, S.; Sun, L. A Comparative Analysis of Edwardsiella tarda-Induced Transcriptome Profiles in RAW264.7 Cells Reveals New Insights into the Strategy of Bacterial Immune Evasion. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Chen, R.; Zhang, L.; Wang, Z.; Yang, D.; Zhang, Y.; Liu, X.; Liu, Q. Balanced role of T3SS and T6SS in contribution to the full virulence of Edwardsiella piscicida. Fish Shellfish Immunol. 2019, 93, 871–878. [Google Scholar] [CrossRef]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef]
- Giacomodonato, M.N.; Uzzau, S.; Bacciu, D.; Caccuri, R.; Sarnacki, S.H.; Rubino, S.; Cerquetti, M.C. SipA, SopA, SopB, SopD and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology 2007, 153, 1221–1228. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Han, F.; Tan, J.; Hou, M.; Zhang, Y.; Yang, D.; Liu, Q. Edwardsiella piscicida Type III Secretion System Effector EseK Inhibits Mitogen-Activated Protein Kinase Phosphorylation and Promotes Bacterial Colonization in Zebrafish Larvae. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.X.; Yu, H.B.; Zheng, J.; Nie, P.; Foster, L.J.; Mok, Y.K.; Finlay, B.B.; Leung, K.Y. EseG, an effector of the type III secretion system of Edwardsiella tarda, triggers microtubule destabilization. Infect. Immun. 2010, 78, 5011–5021. [Google Scholar] [CrossRef] [Green Version]
- Gallique, M.; Bouteiller, M.; Merieau, A. The Type VI Secretion System: A Dynamic System for Bacterial Communication? Front. Microbiol. 2017, 8, 1454. [Google Scholar] [CrossRef]
- Chen, H.; Yang, D.; Han, F.; Tan, J.; Zhang, L.; Xiao, J.; Zhang, Y.; Liu, Q. The Bacterial T6SS Effector EvpP Prevents NLRP3 Inflammasome Activation by Inhibiting the Ca (2+)-Dependent MAPK-Jnk Pathway. Cell Host Microbe 2017, 21, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Tipton, K.A.; Rather, P.N. An ompR-envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter baumannii Strain AB5075. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.H.; Chen, Y.; Kuo, J.T.; Lai, Y.C.; Wu, C.C.; Huang, C.F.; Lin, C.T. Phosphorylated OmpR Is Required for Type 3 Fimbriae Expression in Klebsiella pneumoniae Under Hypertonic Conditions. Front. Microbiol. 2018, 9, 2405. [Google Scholar] [CrossRef] [PubMed]
- Raczkowska, A.; Skorek, K.; Bielecki, J.; Brzostek, K. OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuwenhoek 2011, 99, 381–394. [Google Scholar] [CrossRef] [Green Version]
- Rentschler, A.E.; Lovrich, S.D.; Fitton, R.; Enos-Berlage, J.; Schwan, W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 2013, 159, 316–327. [Google Scholar] [CrossRef] [Green Version]
- Arnold, A. Major Molecular Genetic Drivers in Sporadic Primary Hyperparathyroidism. Trans. Am. Clin. Climatol. Assoc. 2016, 127, 235–244. [Google Scholar] [PubMed]
- Pletzer, D.; Weingart, H. Characterization of AcrD, a resistance-nodulation-cell division-type multidrug efflux pump from the fire blight pathogen Erwinia amylovora. BMC Microbiol. 2014, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Verhamme, D.T.; Postma, P.W.; Crielaard, W.; Hellingwerf, K.J. Cooperativity in signal transfer through the Uhp system of Escherichia coli. J. Bacteriol. 2002, 184, 4205–4210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodorou, E.C.; Theodorou, M.C.; Kyriakidis, D.A. AtoSC two-component system is involved in cPHB biosynthesis through fatty acid metabolism in E. coli. Biochim. Biophys. Acta 2011, 1810, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lin, Y.Y.; Lan, C.Y. The Role of the Two-Component System BaeSR in Disposing Chemicals through Regulating Transporter Systems in Acinetobacter baumannii. PLoS ONE 2015, 10, e0132843. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, G. Iron and Virulence in Francisella tularensis. Front. Cell Infect. Microbiol. 2017, 7, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runci, F.; Gentile, V.; Frangipani, E.; Rampioni, G.; Leoni, L.; Lucidi, M.; Visaggio, D.; Harris, G.; Chen, W.; Stahl, J.; et al. Contribution of Active Iron Uptake to Acinetobacter baumannii Pathogenicity. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [Green Version]
- Elhassanny, A.E.; Anderson, E.S.; Menscher, E.A.; Roop, R.M., 2nd. The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol. Microbiol. 2013, 88, 1070–1082. [Google Scholar] [CrossRef]
- Sit, B.; Crowley, S.M.; Bhullar, K.; Lai, C.C.; Tang, C.; Hooda, Y.; Calmettes, C.; Khambati, H.; Ma, C.; Brumell, J.H.; et al. Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen. PLoS Pathog. 2015, 11, e1005107. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Tung, S.L.; Leung, K.Y. Regulation of a type III and a putative secretion system in Edwardsiella tarda by EsrC is under the control of a two-component system, EsrA-EsrB. Infect. Immun. 2005, 73, 4127–4137. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Wu, Y.; Qiao, H.; Xu, W.; Zhang, Y.; Liu, X.; Wang, Q. YebC controls virulence by activating T3SS gene expression in the pathogen Edwardsiella piscicida. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Li, M.; Chatterjee, C.; Sivaraman, J.; Leung, K.Y.; Mok, Y.K. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J. Biol. Chem. 2010, 285, 38876–38888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Q.; Yang, M.; Xiao, J.; Liu, Q.; Wu, H.; Zhang, Y. QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol. 2011, 30, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Falker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, C.G.; Carneiro, S.M.; Nataro, J.P.; Trabulsi, L.R.; Elias, W.P. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 2003, 226, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Scavone, P.; Iribarnegaray, V.; Caetano, A.L.; Schlapp, G.; Hartel, S.; Zunino, P. Fimbriae have distinguishable roles in Proteus mirabilis biofilm formation. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [Green Version]
- Pakharukova, N.; Tuittila, M.; Paavilainen, S.; Malmi, H.; Parilova, O.; Teneberg, S.; Knight, S.D.; Zavialov, A.V. Structural basis for Acinetobacter baumannii biofilm formation. Proc. Nat. Acad. Sci. USA 2018, 115, 5558–5563. [Google Scholar] [CrossRef] [Green Version]
- Zamani, H.; Salehzadeh, A. Biofilm formation in uropathogenic Escherichia coli: Association with adhesion factor genes. Turk. J. Med. Sci. 2018, 48, 162–167. [Google Scholar] [CrossRef]
- Sarkar, S.; Vagenas, D.; Schembri, M.A.; Totsika, M. Biofilm formation by multidrug resistant Escherichia coli ST131 is dependent on type 1 fimbriae and assay conditions. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; He, T.T.; Li, D.Y.; Liu, L.Y.; Nie, P.; Xie, H.X. The Edwardsiella piscicida Type III Effector EseJ Suppresses Expression of Type 1 Fimbriae, Leading to Decreased Bacterial Adherence to Host Cells. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Hu, Y.H.; Chi, H.; Sun, L. The major fimbrial subunit protein of Edwardsiella tarda: Vaccine potential, adjuvant effect, and involvement in host infection. Fish Shellfish Immunol. 2013, 35, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, M. Strategies to Block Bacterial Pathogenesis by Interference with Motility and Chemotaxis. Curr. Top. Microbiol. Immunol. 2016, 398, 185–205. [Google Scholar] [CrossRef]
- Duan, Q.; Zhou, M.; Zhu, L.; Zhu, G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, M.; Duan, Q.; Zhu, G. Advance on the pathogenicity and immunological application of bacterial flagell—A review. Wei Sheng Wu Xue Bao 2014, 54, 251–260. [Google Scholar] [PubMed]
- Josenhans, C.; Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef]
- Chaban, B.; Hughes, H.V.; Beeby, M. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin. Cell Dev. Biol. 2015, 46, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Swartz, J.R. Functional properties of flagellin as a stimulator of innate immunity. Sci. Rep. 2016, 6, 18379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musser, J.H.; Kreft, A.F.; Bender, R.H.; Kubrak, D.M.; Carlson, R.P.; Chang, J.; Hand, J.M. N-[(arylmethoxy)phenyl] and N-[(arylmethoxy)naphthyl] sulfonamides: Potent orally active leukotriene D4 antagonists of novel structure. J. Med. Chem. 1989, 32, 1176–1183. [Google Scholar] [CrossRef]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, J.B.; Hwang, S.; Baek, S.-W.; Lee, S.; Bang, W.Y.; Moon, K.H. In vitro Edwardsiella piscicida CK108 Transcriptome Profiles with Subinhibitory Concentrations of Phenol and Formalin Reveal New Insights into Bacterial Pathogenesis Mechanisms. Microorganisms 2020, 8, 1068. https://doi.org/10.3390/microorganisms8071068
Yoon JB, Hwang S, Baek S-W, Lee S, Bang WY, Moon KH. In vitro Edwardsiella piscicida CK108 Transcriptome Profiles with Subinhibitory Concentrations of Phenol and Formalin Reveal New Insights into Bacterial Pathogenesis Mechanisms. Microorganisms. 2020; 8(7):1068. https://doi.org/10.3390/microorganisms8071068
Chicago/Turabian StyleYoon, Ju Bin, Sungmin Hwang, Se-Won Baek, Seungki Lee, Woo Young Bang, and Ki Hwan Moon. 2020. "In vitro Edwardsiella piscicida CK108 Transcriptome Profiles with Subinhibitory Concentrations of Phenol and Formalin Reveal New Insights into Bacterial Pathogenesis Mechanisms" Microorganisms 8, no. 7: 1068. https://doi.org/10.3390/microorganisms8071068
APA StyleYoon, J. B., Hwang, S., Baek, S. -W., Lee, S., Bang, W. Y., & Moon, K. H. (2020). In vitro Edwardsiella piscicida CK108 Transcriptome Profiles with Subinhibitory Concentrations of Phenol and Formalin Reveal New Insights into Bacterial Pathogenesis Mechanisms. Microorganisms, 8(7), 1068. https://doi.org/10.3390/microorganisms8071068