A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Signal Preprocessing
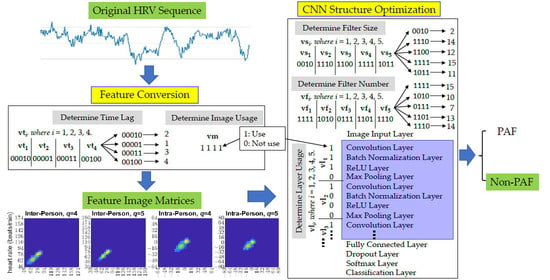
2.3. Feature Matrices Conversion
2.4. Feature Selection and Classification
2.5. Parameter Determinations
3. Results
3.1. Data Split
3.2. The Converted Feature Matrices
3.3. Performance of the Developed PAF Prediction System
4. Discussion
4.1. Comparison with Similar Systems
4.2. Medical Implications from the Features
4.3. Robustness of the System
4.4. Analysis of Misclassifications
4.5. Future Works
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


| No. | Name | Type | Activations | Learnables |
|---|---|---|---|---|
| 1 | imageinput 32 × 32 × 4 images with ‘zerocenter’ normalization | Image Input | 32 × 32 × 4 | - |
| 2 | conv_1 16 3 × 3 × 4 convolutions with stride [1 1] and padding ‘same’ | Convolution | 32 × 32 × 16 | Weights 3 × 3 × 4 × 16 Bias 1 × 1 × 16 |
| 3 | relu_1 ReLU | ReLU | 32 × 32 × 16 | - |
| 4 | conv_2 16 15 × 15 × 16 convolutions with stride [1 1] and padding ‘same’ | Convolution | 32 × 32 × 16 | Weights 15 × 15 × 16 × 16 Bias 1 × 1 × 16 |
| 5 | batchnorm_1 Batch normalization with 16 channels | Batch Normalization | 32 × 32 × 16 | Offset 1 × 1 × 16 Scale 1 × 1 × 16 |
| 6 | conv_3 16 13 × 13 × 16 convolutions with stride [1 1] and padding ‘same’ | Convolution | 32 × 32 × 16 | Weights 13 × 13 × 16 × 16 Bias 1 × 1 × 16 |
| 7 | conv_4 14 16 × 16 × 16 convolutions with stride [1 1] and padding ‘same’ | Convolution | 32 × 32 × 14 | Weights 16 × 16 × 16 × 14 Bias 1 × 1 × 14 |
| 8 | batchnorm_2 Batch normalization with 14 channels | Batch Normalization | 32 × 32 × 14 | Offset 1 × 1 × 14 Scale 1 × 1 × 14 |
| 9 | relu_2 ReLU | ReLU | 32 × 32 × 14 | - |
| 10 | conv_5 15 12 × 12 × 14 convolutions with stride [1 1] and padding ‘same’ | Convolution | 32 × 32 × 15 | Weights 12 × 12 × 14 × 15 Bias 1 × 1 × 15 |
| 11 | batchnorm_3 Batch normalization with 15 channels | Batch Normalization | 32 × 32 × 15 | Offset 1 × 1 × 15 Scale 1 × 1 × 15 |
| 12 | relu_3 ReLU | ReLU | 32 × 32 × 15 | - |
| 13 | fc 2 fully connected layer | Fully Connected | 1 × 1 × 2 | Weights 2 × 15360 Bias 2 × 1 |
| 14 | dropout 50% dropout | Dropout | 1 × 1 × 2 | - |
| 15 | softmax softmax | Softmax | 1 × 1 × 2 | - |
| 16 | classoutput crossentropyex with classes ‘0′ and ‘1′ | Classification Output | - |
References
- Wyndham, C.R. Atrial fibrillation: The most common arrhythmia. Tex. Hear. Inst. J. 2000, 27, 257–267. [Google Scholar]
- Chugh, S.S.; Havmoeller, R.; Narayanan, K.; Singh, D.; Rienstra, M.; Benjamin, E.J.; Gillum, R.F.; Kim, Y.-H.; McAnulty, J.H., Jr.; Zheng, Z.-J.; et al. Worldwide Epidemiology of Atrial Fibrillation: A Global Burden of Disease 2010 Study. Circulation 2014, 129, 837–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart disease and stroke statistics—2019 update: A report from the American heart association. Circulation 2019, 139, e56–e528, Erratum in Circulation 2020, 141, e33. [Google Scholar] [CrossRef] [PubMed]
- Aronow, W.S.; Banach, M. Atrial fibrillation: The new epidemic of the ageing world. J. Atr. Fibrillation 2009, 1, 154. [Google Scholar] [CrossRef]
- Pillarisetti, J.; Patel, A.; Boc, K.; Bommana, S.; Sawers, Y.; Vanga, S.; Sayana, H.; Chen, W.; Nath, J.; Vacek, J. Evolution of paroxysmal atrial fibrillation to persistent or permanent atrial fibrillation: Predictors of progression. J. Atr. Fibrillation 2009, 2, 191. [Google Scholar]
- Peng, C.-K.; Havlin, S.; Hausdorff, J.; Mietus, J.; Stanley, H.; Goldberger, A. Fractal mechanisms and heart rate dynamics: Long-range correlations and their breakdown with disease. J. Electrocardiol. 1995, 28, 59–65. [Google Scholar] [CrossRef]
- Peng, C.-K.; Havlin, S.; Stanley, H.E.; Goldberger, A.L. Quantification of Scaling Exponents and Crossover Phenomena in Nonstationary Heartbeat Time Series. CHAOS 1995, 5, 82–87. [Google Scholar] [CrossRef]
- Thong, T.; McNames, J.; Aboy, M.; Goldstein, B. Prediction of Paroxysmal Atrial Fibrillation by Analysis of Atrial Premature Complexes. IEEE Trans. Biomed. Eng. 2004, 51, 561–569. [Google Scholar] [CrossRef]
- Jalali, A.; Lee, M. Atrial Fibrillation Prediction with Residual Network Using Sensitivity and Orthogonality Constraints. IEEE J. Biomed. Health Informatics 2019, 24, 407–413. [Google Scholar] [CrossRef]
- Sessa, F.; Anna, V.; Messina, G.; Cibelli, G.; Monda, V.; Marsala, G.; Ruberto, M.; Biondi, A.; Cascio, O.; Bertozzi, G.; et al. Heart rate variability as predictive factor for sudden cardiac death. Aging 2018, 10, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Boon, K.; Khalil-Hani, M.; Malarvili, M.; Sia, C. Paroxysmal atrial fibrillation prediction method with shorter HRV sequences. Comput. Methods Programs Biomed. 2016, 134, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Narin, A.; Isler, Y.; Ozer, M.; Perc, M. Early prediction of paroxysmal atrial fibrillation based on short-term heart rate variability. Phys. A Stat. Mech. Its Appl. 2018, 509, 56–65. [Google Scholar] [CrossRef]
- Lynn, K.; Chiang, H. A hybrid two-stage approach for paroxysmal atrial fibrillation prognosis problem. In Proceedings of the Computers in Cardiology, Memphis, TN, USA, 22–25 September 2002. [Google Scholar] [CrossRef]
- Lynn, K.-S. Design of a Novel Paroxysmal Atrial Fibrillation Identification System; Cornell University: Ithaca, NY, USA, 2004. [Google Scholar]
- Acharya, U.R.; Joseph, K.P.; Kannathal, N.; Lim, C.M.; Suri, J.S. Heart rate variability: A review. Med. Biol. Eng. Comput. 2006, 44, 1031–1051. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.E. Long Term Storage: An Experimental Study; Constable & Co., Ltd.: London, UK, 1965. [Google Scholar]
- Fano, U. Ionization Yield of Radiations. II. The Fluctuations of the Number of Ions. Phys. Rev. (Ser. I) 1947, 72, 26–29. [Google Scholar] [CrossRef]
- Allan, D. Statistics of atomic frequency standards. Proc. IEEE 1966, 54, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Lempel, A.; Ziv, J. On the complexity of finite sequences. IEEE Trans. Inf. Theory 1976, 22, 75–81. [Google Scholar] [CrossRef]
- Amar, D.; Roistacher, N.; Zhang, H.; Baum, M.S.; Ginsburg, I.; Steinberg, J.S. Signal-averaged P-wave Duration Does Not Predict Atrial Fibrillation after Thoracic Surgery. J. Am. Soc. Anesthesiol. 1999, 91, 16–23. [Google Scholar] [CrossRef]
- Dilaveris, P.E.; Gialafos, E.J.; Sideris, S.K.; Theopistou, A.M.; Andrikopoulos, G.K.; Kyriakidis, M.; Gialafos, J.E.; Toutouzas, P.K. Simple electrocardiographic markers for the prediction of paroxysmal idiopathic atrial fibrillation. Am. Hear. J. 1998, 135, 733–738. [Google Scholar] [CrossRef]
- Holzfuss, J.; Mayer-Kress, G. An Approach to Error-Estimation in the Application of Dimension Algorithms. In Dimensions and Entropies in Chaotic Systems; Springer: Berlin/Heidelberg, Germany, 1986; pp. 114–122. [Google Scholar] [CrossRef] [Green Version]
- Grassberger, P.; Procaccia, I. Characterization of Strange Attractors. Phys. Rev. Lett. 1983, 50, 346–349. [Google Scholar] [CrossRef]
- DePetrillo, P.B.; Speers, D.; Ruttimann, U.E. Determining the Hurst exponent of fractal time series and its application to electrocardiographic analysis. Comput. Biol. Med. 1999, 29, 393–406. [Google Scholar] [CrossRef]
- Wolf, A.; Swift, J.B.; Swinney, H.L.; Vastano, J.A. Determining Lyapunov exponents from a time series. Phys. D Nonlinear Phenom. 1985, 16, 285–317. [Google Scholar] [CrossRef] [Green Version]
- Eckmann, J.-P.; Ruelle, D. Ergodic theory of chaos and strange attractors. Rev. Mod. Phys. 1985, 57, 617–656. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy: A complexity measure for biological time series data. In Proceedings of the 1991 IEEE Seventeenth Annual Northeast Bioengineering Conference, Hartford, CT, USA, 4–5 April 1991; pp. 35–36. [Google Scholar]
- Moeslund, T.B.; Granum, E. A Survey of Computer Vision-Based Human Motion Capture. Comput. Vis. Image Underst. 2001, 81, 231–268. [Google Scholar] [CrossRef]
- Nassif, A.B.; Shahin, I.; Attili, I.; Azzeh, M.; Shaalan, K. Speech Recognition Using Deep Neural Networks: A Systematic Review. IEEE Access 2019, 7, 19143–19165. [Google Scholar] [CrossRef]
- Kanan, T.; Sadaqa, O.; Aldajeh, A.; Alshwabka, H.; AlZu’bi, S.; Elbes, M.; Hawashin, B.; Alia, M.A. A review of natural language processing and machine learning tools used to analyze arabic social media. In Proceedings of the 2019 IEEE Jordan International Joint Conference on Electrical Engineering and Information Technology (JEEIT), Amman, Jordan, 9–11 April 2019; pp. 622–628. [Google Scholar]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Laguna, P. New Electrocardiographic Signal Processing Techniques: Application to Long-Term Records. PhD. Thesis, Science Faculty, University of Zaragoza, Zaragoza, Spain, 1990. [Google Scholar]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 1985, BME-32, 230–236. [Google Scholar] [CrossRef]
- Goovaerts, H.G.; Ros, H.H.; Akker, T.J.V.D.; Schneider, H. A Digital QRS Detector Based on the Principle of Contour Limiting. IEEE Trans. Biomed. Eng. 1976, 23, 154–160. [Google Scholar] [CrossRef]
- Thakor, N.; Webster, J.G.; Tompkins, W.J. Optimal QRS detector. Med Biol. Eng. Comput. 1983, 21, 343–350. [Google Scholar] [CrossRef]
- Liu, F.; Liu, C.; Jiang, X.; Zhang, Z.; Zhang, Y.; Li, J.; Wei, S. Performance Analysis of Ten Common QRS Detectors on Different ECG Application Cases. J. Healthc. Eng. 2018, 2018, 9050812. [Google Scholar] [CrossRef] [Green Version]
- Brennan, M.; Palaniswami, M.; Kamen, P. Do existing measures of Poincare plot geometry reflect nonlinear features of heart rate variability? IEEE Trans. Biomed. Eng. 2001, 48, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Kamen, P.W.; Krum, H.; Tonkin, A.M. Poincaré Plot of Heart Rate Variability Allows Quantitative Display of Parasympathetic Nervous Activity in Humans. Clin. Sci. 1996, 91, 201–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohebbi, M.; Ghassemian, H. Prediction of paroxysmal atrial fibrillation based on non-linear analysis and spectrum and bispectrum features of the heart rate variability signal. Comput. Methods Programs Biomed. 2012, 105, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.; Jeon, M. Atrial fibrillation detection by heart rate variability in Poincare plot. Biomed. Eng. Online 2009, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsi, A.; Glavin, M.; Jones, E.; Byrne, D. Prediction of paroxysmal atrial fibrillation using new heart rate variability features. Comput. Biol. Med. 2021, 133, 104367. [Google Scholar] [CrossRef]
- Dubin, D. Rapid Interpretation of EKG’s; A Programmed Course; Cover Pub. Co.: Tampa, FL, USA, 1970; 265p. [Google Scholar]
- Hickey, B.; Heneghan, C. Screening for paroxysmal atrial fibrillation using atrial premature contractions and spectral measures. In Proceedings of the Computers in Cardiology, Memphis, TN, USA, 22–25 September 2002; pp. 217–220. [Google Scholar] [CrossRef]
- Chazal, P.d.; Heneghan, C. Automated assessment of atrial fibrillation. In Proceedings of the Computers in Cardiology, Rotterdam, The Netherlands, 23–26 September 2001; Volume 28, pp. 117–120. [Google Scholar]








| Dataset | Non-PAF Sequences (Persons) | PAF Sequences (Persons) |
|---|---|---|
| Training | 444 (37) | 516 (46) |
| Validation | 120 (10) | 120 (10) |
| Testing | 3850 (18) | 1035 (21) |
| Literature | Databases | Data Length (min) | Features | Cross-Validation | Results (%) | ||
|---|---|---|---|---|---|---|---|
| SEN | SPE | ACC | |||||
| Hickey and Henegham [44] | AFPDB | 5 | HRV power spectral density and PACs | 5-fold | 51.0 | 79.0 | 68.0 |
| Chazal and Henegham [45] | AFPDB | 5 | P-wave power spectral density | 5-fold | 81.0 | 69.0 | 75.6 |
| Boon et al. [9] | AFPDB | 5 | Combination of 9 HRV features in time and frequency domains | 10-fold | 86.8 | 88.7 | 87.7 |
| Narin et al. [10] | AFPDB | 5 | Combination of 26 HRV features in time and frequency domains | 10-fold | 92.0 | 88.0 | 90.0 |
| Mendez et al. (This study) | AFPDB * NSRDB * AFDB * | ~5 (400 points) | 32 × 32 × 4 images from extended and discretized Poincaré plot | Single-fold | 80.4 | 89.0 | 87.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendez, M.M.; Hsu, M.-C.; Yuan, J.-T.; Lynn, K.-S. A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System. Appl. Sci. 2022, 12, 2387. https://doi.org/10.3390/app12052387
Mendez MM, Hsu M-C, Yuan J-T, Lynn K-S. A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System. Applied Sciences. 2022; 12(5):2387. https://doi.org/10.3390/app12052387
Chicago/Turabian StyleMendez, Milna Maria, Min-Chia Hsu, Jenq-Tay Yuan, and Ke-Shiuan Lynn. 2022. "A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System" Applied Sciences 12, no. 5: 2387. https://doi.org/10.3390/app12052387
APA StyleMendez, M. M., Hsu, M. -C., Yuan, J. -T., & Lynn, K. -S. (2022). A Heart Rate Variability-Based Paroxysmal Atrial Fibrillation Prediction System. Applied Sciences, 12(5), 2387. https://doi.org/10.3390/app12052387