Sequence-Specific Recognition of Double-Stranded DNA by Peptide Nucleic Acid Forming Double-Duplex Invasion Complex
Abstract
:1. Introduction
2. Double-Stranded DNA Recognition by Peptide Nucleic Acid (PNA)
2.1. Peptide Nucleic Acid (PNA)
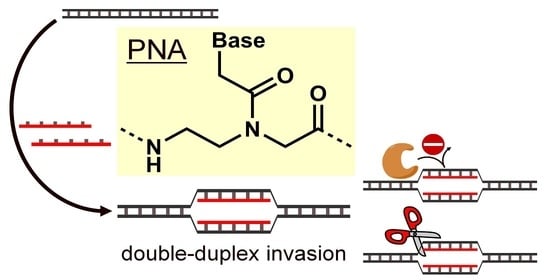
2.2. Invasion Complex Formation by PNA
2.3. Double-Duplex Invasion by Pseudo-Complementary PNAs (pcPNAs)
3. Promotion of Double-Duplex Invasion by Modified PNAs
3.1. Chiral PNAs
3.2. PNA Modified with Nuclear Localization Signal (NLS) Peptide (NLS-PNA)
3.3. Ruthenium-Complex PNA Conjugate (Ru-PNA)
3.4. PNA Containing Cationic Guanine (G+-PNA)
4. Recent Invasion-Related Research
4.1. Development of New Backbone- or Nucleobase-Modified PNAs for DNA Recognition
4.2. Double-Stranded DNA Recognition by Non-PNA-Type Artificial Nucleic Acids
5. Application of PNA Invasion to Biological Research
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by Cys(2)His(2) zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef]
- Jantz, D.; Amann, B.T.; Gatto, G.J.; Berg, J.M. The Design of Functional DNA-Binding Proteins Based on Zinc Finger Domains. Chem. Rev. 2004, 104, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, M.; Negi, S.; Sugiura, Y. Designer zinc finger proteins: Tools for creating artificial DNA-binding functional proteins. Acc. Chem. Res. 2006, 39, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trauger, J.W.; Baird, E.E.; Dervan, P.B. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature 1996, 382, 559–561. [Google Scholar] [CrossRef]
- Dervan, P.B.; Burli, R.W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999, 3, 688–693. [Google Scholar] [CrossRef]
- Dervan, P.B. Molecular recognition of DNA by small molecules. Biorg. Med. Chem. 2001, 9, 2215–2235. [Google Scholar] [CrossRef]
- Dervan, P.B.; Edelson, B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003, 13, 284–299. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Obika, S.; Nanbu, D.; Hari, Y.; Morio, K.-I.; In, Y.; Ishida, T.; Imanishi, T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar] [CrossRef]
- Steffens, R.; Leumann, C.J. Tricyclo-DNA: A Phosphodiester-Backbone Based DNA Analog Exhibiting Strong Complementary Base-Pairing Properties. J. Am. Chem. Soc. 1997, 119, 11548–11549. [Google Scholar] [CrossRef]
- Oka, N.; Wada, T.; Saigo, K. An Oxazaphospholidine Approach for the Stereocontrolled Synthesis of Oligonucleoside Phosphorothioates. J. Am. Chem. Soc. 2003, 125, 8307–8317. [Google Scholar] [CrossRef]
- Zhang, L.; Peritz, A.; Meggers, E. A Simple Glycol Nucleic Acid. J. Am. Chem. Soc. 2005, 127, 4174–4175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, T.S.; Madsen, A.S.; Wengel, J.; Hrdlicka, P.J. Synthesis and Hybridization Studies of 2′-Amino-α-L-LNA and Tetracyclic “Locked LNA”. J. Org. Chem. 2006, 71, 4188–4201. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Wengel, J. Locked vs. unlocked nucleic acids (LNA vs. UNA): Contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 2011, 40, 5680–5689. [Google Scholar] [CrossRef]
- Kashida, H.; Murayama, K.; Toda, T.; Asanuma, H. Control of the Chirality and Helicity of Oligomers of Serinol Nucleic Acid (SNA) by Sequence Design. Angew. Chem. Int. Ed. 2011, 50, 1285–1288. [Google Scholar] [CrossRef]
- Oka, N.; Wada, T. Stereocontrolled synthesis of oligonucleotide analogs containing chiral internucleotidic phosphorus atoms. Chem. Soc. Rev. 2011, 40, 5829–5843. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Yoshimoto, K.; Sisido, M.; Ariga, K. Chemistry Can Make Strict and Fuzzy Controls for Bio-Systems: DNA Nanoarchitectonics and Cell-Macromolecular Nanoarchitectonics. Bull. Chem. Soc. Jpn. 2017, 90, 967–1004. [Google Scholar] [CrossRef] [Green Version]
- Takezawa, Y.; Muller, J.; Shionoya, M. Artificial DNA Base Pairing Mediated by Diverse Metal Ions. Chem. Lett. 2017, 46, 622–633. [Google Scholar] [CrossRef] [Green Version]
- Ukale, D.U.; Lönnberg, T. 2,6-Dimercuriphenol as a Bifacial Dinuclear Organometallic Nucleobase. Angew. Chem. Int. Ed. 2018, 57, 16171–16175. [Google Scholar] [CrossRef]
- Kimoto, M.; Hirao, I. Genetic alphabet expansion technology by creating unnatural base pairs. Chem. Soc. Rev. 2020, 49, 7602–7626. [Google Scholar] [CrossRef]
- Asanuma, H.; Kamiya, Y.; Kashida, H.; Murayama, K. Xeno nucleic acids (XNAs) having non-ribose scaffolds with unique supramolecular properties. Chem. Commun. 2022, 58, 3993–4004. [Google Scholar] [CrossRef]
- Thuong, N.T.; Hélène, C. Sequence-Specific Recognition and Modification of Double-Helical DNA by Oligonucleotides. Angew. Chem. Int. Ed. Engl. 1993, 32, 666–690. [Google Scholar] [CrossRef]
- Praseuth, D.; Guieysse, A.L.; Hélène, C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta Gene Struct. Expr. 1999, 1489, 181–206. [Google Scholar] [CrossRef]
- Fox, K.R. Targeting DNA with triplexes. Curr. Med. Chem. 2000, 7, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Aiba, Y.; Ishizuka, T.; Sumaoka, J. Solid-phase synthesis of pseudo-complementary peptide nucleic acids. Nat. Protoc. 2008, 3, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Demidov, V.V.; Frank-Kamenetskii, M.D. Two sides of the coin: Affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 2004, 29, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E. Peptide Nucleic Acids (PNA) in Chemical Biology and Drug Discovery. Chem. Biodivers. 2010, 7, 786–804. [Google Scholar] [CrossRef]
- Manicardi, A.; Rozzi, A.; Korom, S.; Corradini, R. Building on the peptide nucleic acid (PNA) scaffold: A biomolecular engineering approach. Supramol. Chem. 2017, 29, 784–795. [Google Scholar] [CrossRef]
- Sharma, C.; Awasthi, S.K. Versatility of peptide nucleic acids (PNAs): Role in chemical biology, drug discovery, and origins of life. Chem. Biol. Drug Des. 2017, 89, 16–37. [Google Scholar] [CrossRef]
- Economos, N.G.; Oyaghire, S.; Quijano, E.; Ricciardi, A.S.; Saltzman, W.M.; Glazer, P.M. Peptide Nucleic Acids and Gene Editing: Perspectives on Structure and Repair. Molecules 2020, 25, 735. [Google Scholar] [CrossRef] [Green Version]
- Muangkaew, P.; Vilaivan, T. Modulation of DNA and RNA by PNA. Bioorg. Med. Chem. Lett. 2020, 30, 127064. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Pradhan, B. Evolution of peptide nucleic acid with modifications of its backbone and application in biotechnology. Chem. Biol. Drug Des. 2021, 97, 865–892. [Google Scholar] [CrossRef]
- Liang, X.; Liu, M.; Komiyama, M. Recognition of Target Site in Various Forms of DNA and RNA by Peptide Nucleic Acid (PNA): From Fundamentals to Practical Applications. Bull. Chem. Soc. Jpn. 2021, 94, 1737–1756. [Google Scholar] [CrossRef]
- Lohse, J.; Dahl, O.; Nielsen, P.E. Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 11804–11808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.K.; Orum, H.; Nielsen, P.E.; Norden, B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry 1997, 36, 5072–5077. [Google Scholar] [CrossRef]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchard, O.; Sonnichsen, S.H.; Nielsen, P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Kuwahara, M.; Arimitsu, M.; Sisido, M. Novel peptide nucleic acid that shows high sequence specificity and all-or-none-type hybridization with the complementary DNA. J. Am. Chem. Soc. 1999, 121, 256–257. [Google Scholar] [CrossRef]
- Wada, T.; Minamimoto, N.; Inaki, Y.; Inoue, Y. Peptide Ribonucleic Acids (PRNA). 2. A Novel Strategy for Active Control of DNA Recognition through Borate Ester Formation. J. Am. Chem. Soc. 2000, 122, 6900–6910. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, M.; Du, L.; Fisher, G.W.; Waggoner, A.; Ly, D.H. Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA). J. Am. Chem. Soc. 2003, 125, 6878–6879. [Google Scholar] [CrossRef]
- Myers, M.C.; Witschi, M.A.; Larionova, N.V.; Franck, J.M.; Haynes, R.D.; Hara, T.; Grajkowski, A.; Appella, D.H. A cyclopentane conformational restraint for a peptide nucleic acid: Design, asymmetric synthesis, and improved binding affinity to DNA and RNA. Org. Lett. 2003, 5, 2695–2698. [Google Scholar] [CrossRef]
- Shirude, P.S.; Kumar, V.A.; Ganesh, K.N. (2S,5R/2R,5S)-aminoethylpipecolyl aepip-aegPNA chimera: Synthesis and duplex/triplex stability. Tetrahedron 2004, 60, 9485–9491. [Google Scholar] [CrossRef]
- Govindaraju, T.; Kumar, V.A.; Ganesh, K.N. Synthesis and evaluation of (1S,2R/1R,2S)-aminocyclohexylglycyl PNAs as conformationally preorganized PNA analogues for DNA/RNA recognition. J. Org. Chem. 2004, 69, 1858–1865. [Google Scholar] [CrossRef] [PubMed]
- Cherny, D.Y.; Belotserkovskii, B.P.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchardt, O.; Berg, R.H.; Nielsen, P.E. DNA unwinding upon strand-displacement binding of a thymine-substituted polyamide to double-stranded DNA. Proc. Natl. Acad. Sci. USA 1993, 90, 1667–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egholm, M.; Christensen, L.; Dueholm, K.L.; Buchardt, O.; Coull, J.; Nielsen, P.E. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995, 23, 217–222. [Google Scholar] [CrossRef]
- Griffith, M.C.; Risen, L.M.; Greig, M.J.; Lesnik, E.A.; Sprankle, K.G.; Griffey, R.H.; Kiely, J.S.; Freier, S.M. Single and Bis Peptide Nucleic-Acids as Triplexing Agents—Binding and Stoichiometry. J. Am. Chem. Soc. 1995, 117, 831–832. [Google Scholar] [CrossRef]
- Bentin, T.; Larsen, H.J.; Nielsen, P.E. Combined triplex/duplex invasion of double-stranded DNA by “tail-clamp” peptide nucleic acid. Biochemistry 2003, 42, 13987–13995. [Google Scholar] [CrossRef]
- Kaihatsu, K.; Shah, R.H.; Zhao, X.; Corey, D.R. Extending recognition by peptide nucleic acids (PNAs): Binding to duplex DNA and inhibition of transcription by tail-clamp PNA-peptide conjugates. Biochemistry 2003, 42, 13996–14003. [Google Scholar] [CrossRef]
- Bahal, R.; Ali Mcneer, N.; Quijano, E.; Liu, Y.; Sulkowski, P.; Turchick, A.; Lu, Y.-C.; Bhunia, D.C.; Manna, A.; Greiner, D.L.; et al. In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nat. Commun. 2016, 7, 13304. [Google Scholar] [CrossRef]
- Ricciardi, A.S.; Bahal, R.; Farrelly, J.S.; Quijano, E.; Bianchi, A.H.; Luks, V.L.; Putman, R.; López-Giráldez, F.; Coşkun, S.; Song, E.; et al. In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 2018, 9, 2481. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.E.; Christensen, L. Strand displacement binding of a duplex-forming homopurine PNA to a homopyrimidine duplex DNA target. J. Am. Chem. Soc. 1996, 118, 2287–2288. [Google Scholar] [CrossRef]
- Aiba, Y.; Yamamoto, Y.; Komiyama, M. Activation of double-stranded DNA by one pcPNA strand for its site-selective scission with Ce-IV/EDTA. Chem. Lett. 2007, 36, 780–781. [Google Scholar] [CrossRef]
- Rapireddy, S.; He, G.; Roy, S.; Armitage, B.A.; Ly, D.H. Strand invasion of mixed-sequence B-DNA by acridine-linked, gamma-peptide nucleic acid (gamma-PNA). J. Am. Chem. Soc. 2007, 129, 15596–15600. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N.; Mizuno, Y.; Kunifuda, H.; Matsumura, K.; Komiyama, M. Promotion of Single-Strand Invasion of PNA to Double-Stranded DNA by Pseudo-Complementary Base Pairing. Bull. Chem. Soc. Jpn. 2019, 92, 330–335. [Google Scholar] [CrossRef]
- Haaima, G.; Hansen, H.F.; Christensen, L.; Dahl, O.; Nielsen, P.E. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 1997, 25, 4639–4643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittung, P.; Nielsen, P.E.; Buchardt, O.; Egholm, M.; Norden, B. DNA-like double helix formed by peptide nucleic acid. Nature 1994, 368, 561–563. [Google Scholar] [CrossRef]
- Haaima, G.; Lohse, A.; Buchardt, O.; Nielsen, P.E. Peptide nucleic acids (PNAs) containing thymine monomers derived from chiral amino acids: Hybridization and solubility properties of D-lysine PNA. Angew. Chem. Int. Ed. 1996, 35, 1939–1942. [Google Scholar]
- Sforza, S.; Corradini, R.; Ghirardi, S.; Dossena, A.; Marchelli, R. DNA binding of a D-lysine-based chiral PNA: Direction control and mismatch recognition. Eur. J. Org. Chem. 2000, 2000, 2905–2913. [Google Scholar] [CrossRef]
- Ishizuka, T.; Yoshida, J.; Yamamoto, Y.; Sumaoka, J.; Tedeschi, T.; Corradini, R.; Sforza, S.; Komiyama, M. Chiral introduction of positive charges to PNA for double-duplex invasion to versatile sequences. Nucleic Acids Res. 2008, 36, 1464–1471. [Google Scholar] [CrossRef] [Green Version]
- Englund, E.A.; Appella, D.H. Gamma-substituted peptide nucleic acids constructed from L-lysine are a versatile scaffold for multifunctional display. Angew. Chem. Int. Ed. 2007, 46, 1414–1418. [Google Scholar] [CrossRef]
- Pooga, M.; Soomets, U.; Hallbrink, M.; Valkna, A.; Saar, K.; Rezaei, K.; Kahl, U.; Hao, J.X.; Xu, X.J.; Wiesenfeld-Hallin, Z.; et al. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 1998, 16, 857–861. [Google Scholar] [CrossRef]
- Bendifallah, N.; Rasmussen, F.W.; Zachar, V.; Ebbesen, P.; Nielsen, P.E.; Koppelhus, U. Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA). Bioconjugate Chem. 2006, 17, 750–758. [Google Scholar] [CrossRef]
- Aiba, Y.; Hamano, Y.; Kameshima, W.; Araki, Y.; Wada, T.; Accetta, A.; Sforza, S.; Corradini, R.; Marchelli, R.; Komiyama, M. PNA-NLS conjugates as single-molecular activators of target sites in double-stranded DNA for site-selective scission. Org. Biomol. Chem. 2013, 11, 5233–5238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiba, Y.; Honda, Y.; Komiyama, M. Promotion of double-duplex invasion of peptide nucleic acids through conjugation with nuclear localization signal peptide. Chem. Eur. J. 2015, 21, 4021–4026. [Google Scholar] [CrossRef] [PubMed]
- Aiba, Y.; Urbina, G.; Shibata, M.; Shoji, O. Investigation of the Characteristics of NLS-PNA: Influence of NLS Location on Invasion Efficiency. Appl. Sci. 2020, 10, 8663. [Google Scholar] [CrossRef]
- Hibino, M.; Aiba, Y.; Watanabe, Y.; Shoji, O. Peptide Nucleic Acid Conjugated with Ruthenium-Complex Stabilizing Double-Duplex Invasion Complex Even under Physiological Conditions. ChemBioChem 2018, 19, 1601–1604. [Google Scholar] [CrossRef] [PubMed]
- Hibino, M.; Aiba, Y.; Shoji, O. Cationic guanine: Positively charged nucleobase with improved DNA affinity inhibits self-duplex formation. Chem. Commun. 2020, 56, 2546–2549. [Google Scholar] [CrossRef] [PubMed]
- Hendler, S.S.; Fuerer, E.; Srinivasan, P.R. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerases. Biochemistry 1970, 9, 4141–4153. [Google Scholar] [CrossRef]
- Vilaivan, T.; Srisuwannaket, C. Hybridization of Pyrrolidinyl Peptide Nucleic Acids and DNA: Selectivity, Base-Pairing Specificity, and Direction of Binding. Org. Lett. 2006, 8, 1897–1900. [Google Scholar] [CrossRef] [PubMed]
- Mansawat, W.; Vilaivan, C.; Balázs, Á.; Aitken, D.J.; Vilaivan, T. Pyrrolidinyl Peptide Nucleic Acid Homologues: Effect of Ring Size on Hybridization Properties. Org. Lett. 2012, 14, 1440–1443. [Google Scholar] [CrossRef]
- Vilaivan, T. Pyrrolidinyl PNA with alpha/beta-Dipeptide Backbone: From Development to Applications. Acc. Chem Res. 2015, 48, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Yotapan, N.; Nim-anussornkul, D.; Vilaivan, T. Pyrrolidinyl peptide nucleic acid terminally labeled with fluorophore and end-stacking quencher as a probe for highly specific DNA sequence discrimination. Tetrahedron 2016, 72, 7992–7999. [Google Scholar] [CrossRef]
- Gupta, M.K.; Madhanagopal, B.R.; Datta, D.; Ganesh, K.N. Structural Design and Synthesis of Bimodal PNA That Simultaneously Binds Two Complementary DNAs To Form Fused Double Duplexes. Org. Lett. 2020, 22, 5255–5260. [Google Scholar] [CrossRef] [PubMed]
- Bhingardeve, P.; Madhanagopal, B.R.; Ganesh, K.N. Cγ(S/R)-Bimodal Peptide Nucleic Acids (Cγ-bm-PNA) Form Coupled Double Duplexes by Synchronous Binding to Two Complementary DNA Strands. J. Org. Chem. 2020, 85, 13680–13693. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Madhanagopal, B.R.; Ganesh, K.N. Peptide Nucleic Acid with Double Face: Homothymine–Homocytosine Bimodal Cα-PNA (bm-Cα-PNA) Forms a Double Duplex of the bm-PNA2:DNA Triplex. J. Org. Chem. 2021, 86, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Bhingardeve, P.; Jain, P.; Ganesh, K.N. Molecular Assembly of Triplex of Duplexes from Homothyminyl-Homocytosinyl Cγ(S/R)-Bimodal Peptide Nucleic Acids with dA8/dG6 and the Cell Permeability of Bimodal Peptide Nucleic Acids. ACS Omega 2021, 6, 19757–19770. [Google Scholar] [CrossRef] [PubMed]
- Thadke, S.A.; Hridya, V.M.; Perera, J.D.R.; Gil, R.R.; Mukherjee, A.; Ly, D.H. Shape selective bifacial recognition of double helical DNA. Commun. Chem. 2018, 1, 79. [Google Scholar] [CrossRef] [Green Version]
- Guenther, D.C.; Anderson, G.H.; Karmakar, S.; Anderson, B.A.; Didion, B.A.; Guo, W.; Verstegen, J.P.; Hrdlicka, P.J. Invader probes: Harnessing the energy of intercalation to facilitate recognition of chromosomal DNA for diagnostic applications. Chem. Sci. 2015, 6, 5006–5015. [Google Scholar] [CrossRef] [Green Version]
- Guenther, D.C.; Karmakar, S.; Hrdlicka, P.J. Bulged Invader probes: Activated duplexes for mixed-sequence dsDNA recognition with improved thermodynamic and kinetic profiles. Chem. Commun. 2015, 51, 15051–15054. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.A.; Hrdlicka, P.J. Merging Two Strategies for Mixed-Sequence Recognition of Double-Stranded DNA: Pseudocomplementary Invader Probes. J. Org. Chem. 2016, 81, 3335–3346. [Google Scholar] [CrossRef] [Green Version]
- Karmakar, S.; Horrocks, T.; Gibbons, B.C.; Guenther, D.C.; Emehiser, R.; Hrdlicka, P.J. Synthesis and biophysical characterization of oligonucleotides modified with O2′-alkylated RNA monomers featuring substituted pyrene moieties. Org. Biomol. Chem. 2019, 17, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.P.; Vukelich, P.; Guenther, D.C.; Karmakar, S.; Hrdlicka, P.J. Recognition of double-stranded DNA using LNA-modified toehold Invader probes. Org. Biomol. Chem. 2021, 19, 9276–9290. [Google Scholar] [CrossRef]
- Adhikari, S.P.; Karmakar, S.; Hrdlicka, P.J. Nicked Invader probes: Multistranded and sequence-unrestricted recognition of double-stranded DNA. Org. Biomol. Chem. 2022, 20, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Niwa, R.; Akahane, M.; Murayama, K.; Kashida, H.; Kamiya, Y. Strand-invading linear probe combined with unmodified PNA. Bioorg. Med. Chem. 2016, 24, 4129–4137. [Google Scholar] [CrossRef]
- Asanuma, H.; Akahane, M.; Kondo, N.; Osawa, T.; Kato, T.; Kashida, H. Quencher-free linear probe with multiple fluorophores on an acyclic scaffold. Chem. Sci. 2012, 3, 3165–3169. [Google Scholar] [CrossRef]
- Asanuma, H.; Akahane, M.; Niwa, R.; Kashida, H.; Kamiya, Y. Highly sensitive and robust linear probe for detection of mRNA in cells. Angew. Chem. Int. Ed. 2015, 54, 4315–4319. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kawabata, H.; Fujimoto, K. Double duplex invasion of DNA induced by ultrafast photo-cross-linking using 3-cyanovinylcarbazole for antigene methods. Chem. Commun. 2017, 53, 7616–7619. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Fujimoto, K. Ultrafast Reversible Photo-Cross-Linking Reaction: Toward in Situ DNA Manipulation. Org. Lett. 2008, 10, 3227–3230. [Google Scholar] [CrossRef]
- Fujimoto, K.; Yamada, A.; Yoshimura, Y.; Tsukaguchi, T.; Sakamoto, T. Details of the Ultrafast DNA Photo-Cross-Linking Reaction of 3-Cyanovinylcarbazole Nucleoside: Cis–Trans Isomeric Effect and the Application for SNP-Based Genotyping. J. Am. Chem. Soc. 2013, 135, 16161–16167. [Google Scholar] [CrossRef]
- Fujimoto, K.; Hirano, A.; Watanabe, Y.; Shimabara, A.; Nakamura, S. The Inhibition Effect of Photo-Cross-Linking between Probes in Photo-Induced Double Duplex Invasion DNA. ChemBioChem 2021, 22, 3402–3405. [Google Scholar] [CrossRef]
- Izvolsky, K.I.; Demidov, V.V.; Nielsen, P.E.; Frank-Kamenetskii, M.D. Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry 2000, 39, 10908–10913. [Google Scholar] [CrossRef]
- Protozanova, E.; Demidov, V.V.; Nielsen, P.E.; Frank-Kamenetskii, M.D. Pseudocomplementary PNAs as selective modifiers of protein activity on duplex DNA: The case of type IIs restriction enzymes. Nucleic Acids Res. 2003, 31, 3929–3935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollegaard, N.E.; Buchardt, O.; Egholm, M.; Nielsen, P.E. Peptide nucleic acid-DNA strand displacement loops as artificial transcription promoters. Proc. Natl. Acad. Sci. USA 1994, 91, 3892–3895. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Jing, K.; Balczon, R.; Xu, X. Defining the peptide nucleic acids (PNA) length requirement for PNA binding-induced transcription and gene expression. J. Mol. Biol. 2001, 313, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, M.; Aiba, Y.; Yamamoto, Y.; Sumaoka, J. Artificial restriction DNA cutter for site-selective scission of double-stranded DNA with tunable scission site and specificity. Nat. Protoc. 2008, 3, 655–662. [Google Scholar] [CrossRef]
- Lyu, M.; Kong, L.; Yang, Z.; Wu, Y.; McGhee, C.E.; Lu, Y. PNA-Assisted DNAzymes to Cleave Double-Stranded DNA for Genetic Engineering with High Sequence Fidelity. J. Am. Chem. Soc. 2021, 143, 9724–9728. [Google Scholar] [CrossRef] [PubMed]
- Faruqi, A.F.; Egholm, M.; Glazer, P.M. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc. Natl. Acad. Sci. USA 1998, 95, 1398–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.H.; Nielsen, P.E.; Glazer, P.M. Site-directed gene mutation at mixed sequence targets by psoralen-conjugated pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2007, 35, 7604–7613. [Google Scholar] [CrossRef] [PubMed]
- Lonkar, P.; Kim, K.H.; Kuan, J.Y.; Chin, J.Y.; Rogers, F.A.; Knauert, M.P.; Kole, R.; Nielsen, P.E.; Glazer, P.M. Targeted correction of a thalassemia-associated β-globin mutation induced by pseudo-complementary peptide nucleic acids. Nucleic Acids Res. 2009, 37, 3635–3644. [Google Scholar] [CrossRef] [PubMed] [Green Version]















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiba, Y.; Shibata, M.; Shoji, O. Sequence-Specific Recognition of Double-Stranded DNA by Peptide Nucleic Acid Forming Double-Duplex Invasion Complex. Appl. Sci. 2022, 12, 3677. https://doi.org/10.3390/app12073677
Aiba Y, Shibata M, Shoji O. Sequence-Specific Recognition of Double-Stranded DNA by Peptide Nucleic Acid Forming Double-Duplex Invasion Complex. Applied Sciences. 2022; 12(7):3677. https://doi.org/10.3390/app12073677
Chicago/Turabian StyleAiba, Yuichiro, Masanari Shibata, and Osami Shoji. 2022. "Sequence-Specific Recognition of Double-Stranded DNA by Peptide Nucleic Acid Forming Double-Duplex Invasion Complex" Applied Sciences 12, no. 7: 3677. https://doi.org/10.3390/app12073677
APA StyleAiba, Y., Shibata, M., & Shoji, O. (2022). Sequence-Specific Recognition of Double-Stranded DNA by Peptide Nucleic Acid Forming Double-Duplex Invasion Complex. Applied Sciences, 12(7), 3677. https://doi.org/10.3390/app12073677