Growth Identification of Aspergillus flavus and Aspergillus parasiticus by Visible/Near-Infrared Hyperspectral Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Image Acquisition and Calibration
2.3. Data Analysis Method
2.3.1. Explanatory Method
2.3.2. Model Development Method
3. Result and Discussion
3.1. Hyperspectral Image Preprocessing
3.2. Identification of Fungal Growth Days by Band Math
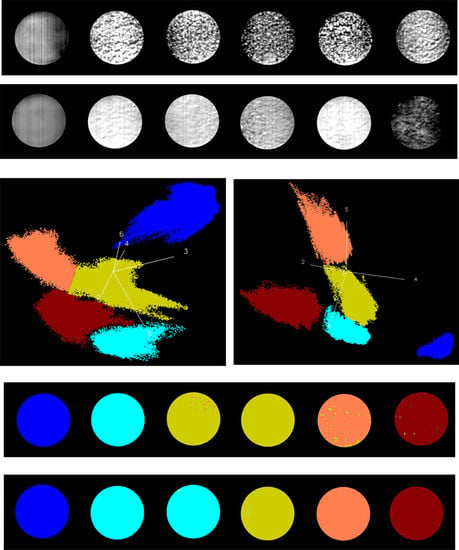
3.3. Identification of Fungal Growth Days by Explanatory Principle Component Analysis (PCA)
3.4. Identification of Fungal Growth Days by Support Vector Machine (SVM)
3.4.1. Full-Spectrum SVM Models
3.4.2. Optimal Wavelengths SVM Models
4. Conclusions
- Band math and PCA on hyperspectral image can be used to classify fungi with distinct characteristics. A band ratio of 446 nm and 460 nm classified A. flavus and A. parasiticus on day 1. The image at the band of 520 nm classified A. parasiticus on day 6. The 5-dimensional score plot of PC2 to PC6 gave an indication of clusters fungi in the same incubation time, with the exception of A. flavus on day 3 and day 4 and A. parasiticus on day 2 and day 3.
- The SVM model that is based on full spectrum data can be used to classify fungal growth time. On the full spectral, PC2 to PC6 2–6 and the SVM method could be used to classify fungi with different incubation times. The overall classification accuracies were 92.59% and 100% for A. flavus and A. parasiticus, respectively.
- Optimal wavelengths selected by the CARS method can be used to a build an optimized SVM model for identifying the growth of A. flavus and A. parasiticus. Nine (402, 442, 487, 502, 524, 553, 646, 671, and 760 nm) and seven (461, 538, 542, 742, 753, 756, and 919 nm) were selected for A. flavus and A. parasiticus. The accuracies of the optimal wavelengths SVM models were 83.33% for A. flavus and 98.15% for A. parasiticus. The predicted map indicated that the predicted fungal growth days were basically consistent with the actual situation.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gourama, H.; Bullerman, L.B. Aspergillus flavus and Aspergillus parasiticus: Aflatoxigenic fungi of concern in foods and feeds: A Review. J. Food Prot. 1995, 58, 1395–1404. [Google Scholar] [CrossRef]
- Perrone, G.; Susca, A.; Cozzi, G.; Ehrlich, K.; Varga, J.; Frisvad, J.C.; Meijer, M.; Noonim, P.; Mahakarnchanakul, W.; Samson, R.A. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 2007, 59, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Kosegarten, C.E.; Ramírez-Corona, N.; Mani-López, E.; Palou, E.; López-Malo, A. Description of Aspergillus flavus growth under the influence of different factors (water activity, incubation temperature, protein and fat concentration, pH, and cinnamon essential oil concentration) by kinetic, probability of growth, and time-to-detection models. Int. J. Food Microbiol. 2017, 240, 115–123. [Google Scholar] [PubMed]
- Bullerman, L.B. Significance of Mycotoxins to Food Safety and Human Health. J. Food Prot. 1979, 42, 65–86. [Google Scholar] [CrossRef]
- Mandeel, Q.A. Fungal contamination of some imported spices. Mycopathologia 2005, 159, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lu, Z.; Cao, L.; Pang, H.; Zhang, Q.; Fu, Y.; Xiong, Y.; Li, Y.; Wang, X.; Wang, J.; Ying, Y.; Li, Y. In-field detection of multiple pathogenic bacteria in food products using a portable fluorescent biosensing system. Food Control 2017, 75, 21–28. [Google Scholar] [CrossRef]
- Sun, Y.; Gu, X.; Wang, Z.; Huang, Y.; Wei, Y.; Zhang, M.; Tu, K.; Pan, L. Growth simulation and discrimination of Botrytis cinerea, Rhizopus stolonifer and Colletotrichum acutatum using hyperspectral reflectance imaging. PLoS ONE 2015, 10, e0143400. [Google Scholar] [CrossRef] [PubMed]
- Matcham, S.E.; Jordan, B.R.; Wood, D.A. Estimation of fungal biomass in a solid substrate by three independent methods. Appl. Microbiol. Biotechnol. 1985, 21, 108–112. [Google Scholar] [CrossRef]
- Edelstein, L.; Segel, L.A. Growth and metabolism in mycelial fungi. J. Theor. Biol. 1983, 104, 187–210. [Google Scholar] [CrossRef]
- Yoon, S.C.; Lawrence, K.C.; Siragusa, G.R.; Line, J.E.; Park, B.; Feldner, P.W. Hyperspectral reflectance imaging for detecting a foodborne pathogen: Campylobacter. Trans. ASABE 2009, 52, 651–662. [Google Scholar] [CrossRef]
- Kammies, T.-L.; Manley, M.; Gouws, P.A.; Williams, P.J. Differentiation of foodborne bacteria using NIR hyperspectral imaging and multivariate data analysis. Appl. Microbiol. Biotechnol. 2016, 100, 9305–9320. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pu, H.; Sun, D.-W.; Liu, D.; Wang, Q.; Xiong, Z. Application of hyperspectral imaging for prediction of textural properties of maize seeds with different storage periods. Food Anal. Methods 2015, 8, 1535–1545. [Google Scholar] [CrossRef]
- Yoon, S.C.; Windham, W.R.; Ladely, S.; Heitschmidt, G.W.; Lawrence, K.C.; Park, B.; Narang, N.; Cray, W.C. Differentiation of big-six non-O157 Shiga-toxin producing Escherichia coli (STEC) on spread plates of mixed cultures using hyperspectral imaging. J. Food Meas. Charact. 2013, 7, 47–59. [Google Scholar] [CrossRef]
- Serranti, S.; Cesare, D.; Bonifazi, G. The development of a hyperspectral imaging method for the detection of Fusarium-damaged, yellow berry and vitreous Italian durum wheat kernels. Biosyst. Eng. 2013, 115, 20–30. [Google Scholar] [CrossRef]
- Del Fiore, A.; Reverberi, M.; Ricelli, A.; Pinzari, F.; Serranti, S.; Fabbri, A.A.; Bonifazi, G.; Fanelli, C. Early detection of toxigenic fungi on maize by hyperspectral imaging analysis. Int. J. Food Microbiol. 2010, 144, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Falade, T.; Sultanbawa, Y.; Fletcher, M.T.; Fox, G. Near Infrared Spectrometry for Rapid Non-Invasive Modelling of Aspergillus-Contaminated Maturing Kernels of Maize (Zea mays L.). Agriculture 2017, 7, 77. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, W.; Chu, X.; Li, C.; Kimuli, D. Early detection of Aspergillus parasiticus infection in maize kernels using near-infrared hyperspectral imaging and multivariate data analysis. Appl. Sci. 2017, 7, 90. [Google Scholar] [CrossRef]
- Yao, H.; Hruska, Z.; Kincaid, R.; Brown, R.L.; Cleveland, T.E. Differentiation of toxigenic fungi using hyperspectral imagery. Sens. Instrum. Food Qual. Saf. 2008, 2, 215. [Google Scholar] [CrossRef]
- Jin, J.; Tang, L.; Hruska, Z.; Yao, H. Classification of toxigenic and atoxigenic strains of Aspergillus flavus with hyperspectral imaging. Comput. Electron. Agric. 2009, 69, 158–164. [Google Scholar] [CrossRef]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Near-infrared (NIR) hyperspectral imaging and multivariate image analysis to study growth characteristics and differences between species and strains of members of the genus Fusarium. Anal. Bioanal. Chem. 2012, 404, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Growth characteristics of three Fusarium species evaluated by near-infrared hyperspectral imaging and multivariate image analysis. Appl. Microbiol. Biotechnol. 2012, 96, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Dégardin, K.; Guillemain, A.; Guerreiro, N.V.; Roggo, Y. Near infrared spectroscopy for counterfeit detection using a large database of pharmaceutical tablets. J. Pharm. Biomed. Anal. 2016, 128, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Yoon, S.-C.; Zhuang, H.; Wang, W.; Li, C. Prediction of pH of fresh chicken breast fillets by VNIR hyperspectral imaging. J. Food Eng. 2017, 208, 57–65. [Google Scholar] [CrossRef]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Investigation of fungal development in maize kernels using NIR hyperspectral imaging and multivariate data analysis. J. Cereal Sci. 2012, 55, 272–278. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; ElMasry, G.; Sun, D.-W.; Allen, P. Application of NIR hyperspectral imaging for discrimination of lamb muscles. J. Food Eng. 2011, 104, 332–340. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Huang, W. Detection of early bruises on peaches (Amygdalus persica L.) using hyperspectral imaging coupled with improved watershed segmentation algorithm. Postharvest Biol. Technol. 2018, 135, 104–113. [Google Scholar] [CrossRef]
- Fan, S.; Huang, W.; Guo, Z.; Zhang, B.; Zhao, C. Prediction of soluble solids content and firmness of pears using hyperspectral reflectance imaging. Food Anal. Methods 2015, 8, 1936–1946. [Google Scholar] [CrossRef]
- He, H.-J.; Sun, D.-W.; Wu, D. Rapid and real-time prediction of lactic acid bacteria (LAB) in farmed salmon flesh using near-infrared (NIR) hyperspectral imaging combined with chemometric analysis. Food Res. Int. 2014, 62, 476–483. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.-W. Potential of time series-hyperspectral imaging (TS-HSI) for non-invasive determination of microbial spoilage of salmon flesh. Talanta 2013, 111, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Fernández-Ibañez, V.; Soldado, A.; Martínez-Fernández, A.; De la Roza-Delgado, B. Application of near infrared spectroscopy for rapid detection of aflatoxin B1 in maize and barley as analytical quality assessment. Food Chem. 2009, 113, 629–634. [Google Scholar] [CrossRef]











© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, X.; Wang, W.; Ni, X.; Zheng, H.; Zhao, X.; Zhang, R.; Li, Y. Growth Identification of Aspergillus flavus and Aspergillus parasiticus by Visible/Near-Infrared Hyperspectral Imaging. Appl. Sci. 2018, 8, 513. https://doi.org/10.3390/app8040513
Chu X, Wang W, Ni X, Zheng H, Zhao X, Zhang R, Li Y. Growth Identification of Aspergillus flavus and Aspergillus parasiticus by Visible/Near-Infrared Hyperspectral Imaging. Applied Sciences. 2018; 8(4):513. https://doi.org/10.3390/app8040513
Chicago/Turabian StyleChu, Xuan, Wei Wang, Xinzhi Ni, Haitao Zheng, Xin Zhao, Ren Zhang, and Yufeng Li. 2018. "Growth Identification of Aspergillus flavus and Aspergillus parasiticus by Visible/Near-Infrared Hyperspectral Imaging" Applied Sciences 8, no. 4: 513. https://doi.org/10.3390/app8040513
APA StyleChu, X., Wang, W., Ni, X., Zheng, H., Zhao, X., Zhang, R., & Li, Y. (2018). Growth Identification of Aspergillus flavus and Aspergillus parasiticus by Visible/Near-Infrared Hyperspectral Imaging. Applied Sciences, 8(4), 513. https://doi.org/10.3390/app8040513